Department of Integrated Analytics
We deal with multidimensional and ultra-heterogeneous biological big data, which is a combination of omics data such as genome, epigenome, transcriptome, proteome, and metabolome, and human body composition values, medical interviews, blood test values, blood gas analysis values, ECG, ventilator data, and so on. Through the analysis using bioinformatics, statistical science, and machine learning (Bayesian theory, deep learning, etc.) on advanced information processing technologies such as supercomputers and GPUs, we aim to reveal new medical and life science knowledge and to elucidate entire biological systems. Beyond understanding and applying existing bioinformatics methodologies, we’re also developing new mathematical methods to analyze continuously emerging new life science data, and multi-dimensional and ultra-heterogeneous biological data. Through these methodologies and a broad understanding of medicine and life science, we are able to analyze biological systems, elucidate genomic and epigenomic causes of disease, predict patient prognosis and future conditions for acute and chronic diseases, and formulate therapeutic strategies.
Member
Satoru Miyano
Specially Appointed Professor
Takanori Hasegawa
Associate Professor
Hiroko Tanaka
Assistant Professor
Seishi Okamoto
(Fujitsu Ltd.)
Visiting Professor
Koji Maruhashi
(Fujitsu Ltd.)
Visiting Professor
Project
Human Internal state prediction using mathematical modeling
Disease states are caused by the complex interactions of multidimensional and heterogeneous factors such as genes and metabolites in the body, and bacterial flora in the intestines and skin. On the other hand, a healthy state is one in which these factors are balanced within the organism. Our team combines omics data such as genome, transcriptome, proteome, and large-scale patient data obtained through clinical and health examinations with mathematical modeling to clarify the complex relationships among them and elucida the internal states of healthy and diseased individuals. This enables us to determine the causative factors of diseases, predict the severity and prognosis of each patient, and propose treatment strategies for them. In addition, even if we have only partial medical information, such as body composition values and medical interview items, these technologies will estimate the probability of disease through the prediction of blood test values and to identify the part of abnormal states.
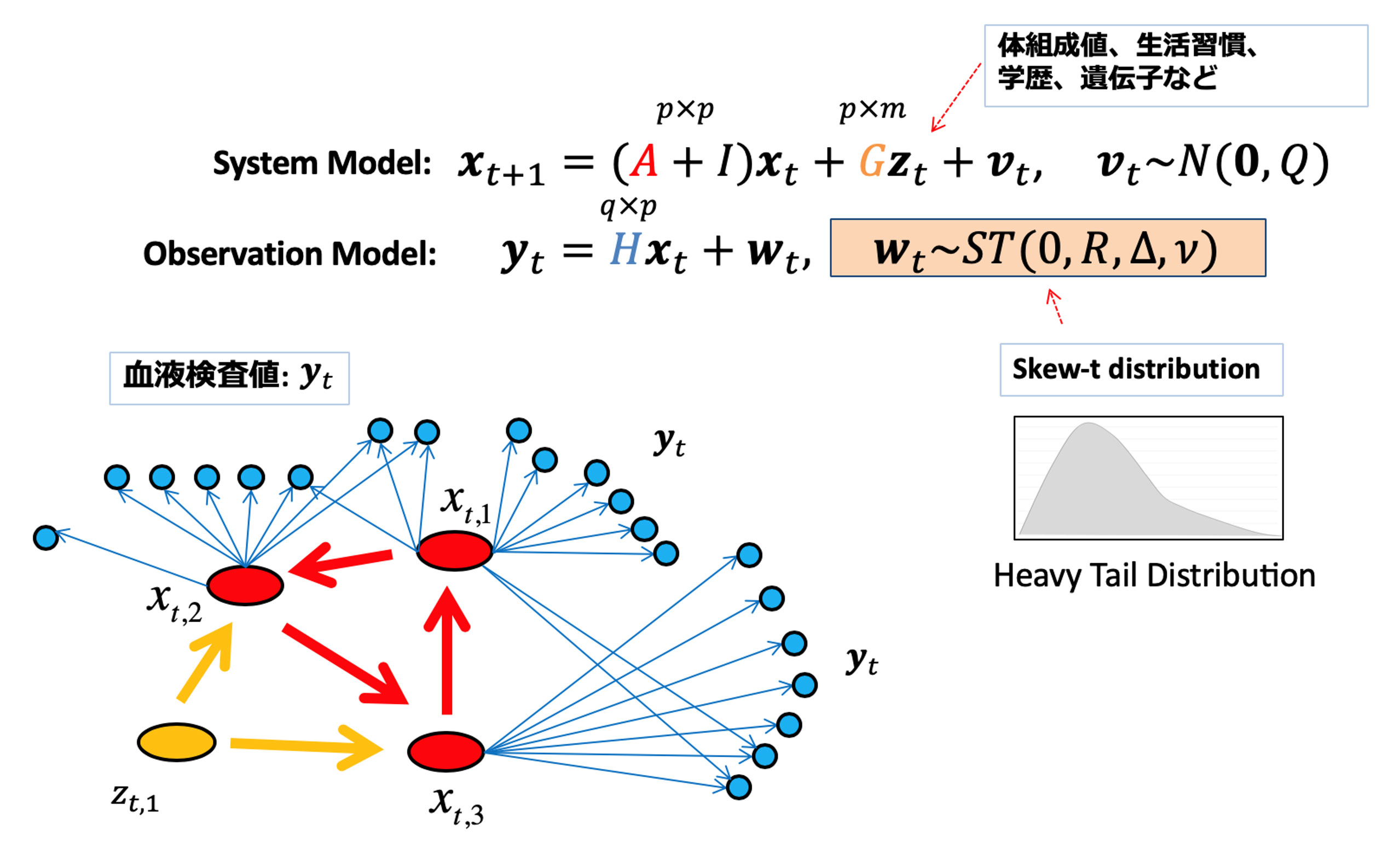
Mathematical modeling of the effects of lifestyle, social status, and genetic factors on blood test values using a state-space model.
Development of neoantigen prediction methods to target tumor cells and inflammation-inducing cells such as senescent cells
With the development of next-generation sequencing, whole genome/exome sequencing has become a standard analytical method in cancer research. For many years, our team has been working to elucidate the pathogenesis of cancer by mathematically analyzing sequencing results. As part of this research, we have developed analytical methods to predict neoantigens based on mutations, analyzed the role of neoantigens in the cancer immune system, and evaluated immune pathology in tumors. In particular, recent studies have shown the possibility of eliminating not only tumor cells but also (partially) senescent cells by using T cells designed to target senescent cells and checkpoint inhibitors. We are developing mathematical analysis methods to assess and elucidate immune system at the cellular level and to predict appropriate neoantigens with the goal of controlling immune system and appropriately regulating these cells.
Large-scale data analysis and artificial intelligence techniques to elucidate the origin and diversity of cancer
Large-scale data analysis and artificial intelligence technology have elucidated the origin and diversity of cancerCancer research over the years. They have revealed that genes involved in invasion and epithelial-mesenchymal transition (EMT) are deeply involved in cancer metastasis. In order to elucidate the mechanisms of invasion and EMT, it is not enough to simply identify the genes involved; we need to know what kind of network is formed among the related genes, which network plays a particularly important role in invasion and EMT within that network structure, and which network is the most important one. Our team utilized the supercomputer system at the Human Genome Center of the Institute of Medical Science, the University of Tokyo, and the K and Fugaku supercomputers to run a program called network profiling, which can estimate networks that are likely to be associated with metastasis or recurrence of cancer cells. We’re extracting the network structure formed by genes that are highly likely to be associated with metastasis and recurrence of cancer cells, and to elucidate the mechanisms of metastasis and recurrence that cannot be determined at the individual gene level, as well as to discover new mechanisms.
Establishment of an integrated analysis platform for M&D data science center and promotion of collaborative research systems within and outside of Science Tokyo
Medical and dental research is entering a new era in terms of both quality and quantity of information. In the analysis of health conditions and diseases, it has become common to obtain large amounts of biomedical information through, e.g., whole genome and single cell analysis, and the integrated analysis of such information can generate new findings and therapeutic strategies for diseases. Our team, Integrated Analytics Division, consists of research members with many years of experience in such large-scale data analysis, and promotes biomedical research not only within Tokyo Medical and Dental University, but also in collaboration with many external research institutions, including the University of Tokyo, Kobe University, and Kyushu University. Our activities include whole genome analysis in the COVID19 Task Force, studies to predict the severity, mortality, and prognosis of acute and chronic diseases including COVID19 patients in ICU, germline and tumor genome analysis (whole genome, exome, transcriptome, single cell analysis, cfDNA analysis, proteome analysis, metabolome analysis, etc.), and drug response prediction. In addition, such as a comprehensive agreement has been concluded with the supercomputer SHIROKANE at the Institute of Medical Science, the University of Tokyo, we will practice integrated analysis of Science Tokyo and promote information education and the construction of an information infrastructure necessary for integrated analysis.
Research Topic
- Human internal state prediction using mathematical modeling
- Prediction of disease incidence and severity using health examination and electronic health record
- Mathematical modeling for the tumor and senescent cells immune system analysis
- Large-scale data analysis and artificial intelligence techniques to elucidate the origin and diversity of cancer
- Large-scale multi-omics data analysis for healthy and diseases persons
Recent Works
- Noguchi R, Yamaguchi K, Yano H, Gohda Y, Kiyomatsu T, Ota Y, Igari T, Takahashi N, Ohsugi T, Takane K, Ikenoue T, Niida A, Shimizu E, Yamaguchi R, Miyano S, Imoto S, Furukawa Y. Cell of origin and expression profiles of pseudomyxoma peritonei derived from the appendix. Pathol Res Pract. 2025 Feb;266:155776. doi: 10.1016/j.prp.2024.155776. Epub 2024 Dec 21. PMID: 39742833 [download]
- Okuda R, Ochi Y, Saiki R, Yamanaka T, Terao C, Yoshizato T, Nakagawa MM, Zhao L, Ohyashiki K, Hiramoto N, Sanada M, Handa H, Kasahara S, Miyazaki Y, Sezaki N, Shih LY, Kern W, Kanemura N, Kitano T, Imashuku S, Watanabe M, Creignou M, Chonabayashi K, Usuki K, Ishikawa T, Gotoh A, Atsuta Y, Shiraishi Y, Mitani K, Chiba S, K, Yoshida Y, Makishima H, Nannya Y, Ogawa S. Genetic analysis of myeloid neoplasms with der(1;7)(q10;p10). Leukemia. 2025 Mar;39(3):760-764. doi: 10.1038/s41375-024-02494-2. Epub 2024 Dec 23. PMID: 39715854 [download]
- Ozawa T, Chubachi S, Namkoong H, Nemoto S, Ikegami R, Asakura T, Tanaka H, Lee H, Fukushima T, Azekawa S, Otake S, Nakagawara K, Watase M, Masaki K, Kamata H, Harada N, Ueda T, Ueda S, Ishiguro T, Arimura K, Saito F, Yoshiyama T, Nakano Y, Muto Y, Suzuki Y, Edahiro R, Murakami K, Sato Y, Okada Y, Koike R, Ishii M, Hasegawa N, Kitagawa Y, Tokunaga K, Kimura A, Miyano S, Ogawa S, Kanai T, Fukunaga K, Imoto S. Predicting coronavirus disease 2019 severity using explainable artificial intelligence techniques. Sci Rep. 2025 Mar 19;15(1):9459. doi: 10.1038/s41598-025-85733-5. PMID: 40108236 [download]
- Park H, Miyano S. Breaking the complexity of cancer using computational transcriptomic network biology. Wiley Interdisciplinary Reviews: Computational Statistics. 2025 April 4; 17:e70020. doi: https://doi.org/10.1002/wics.70020 [download]
- Park H, Miyano S. Computational network biology analysis revealed COVID-19 severity markers: Molecular interplay between HLA-II with CIITA. PLoS One. 2025 Mar 31;20(3):e0319205. doi: 10.1371/journal.pone.0319205. eCollection 2025. PMID: 40163554 [download]
- Park H, Miyano S. Network-based multi-class classifier to identify optimized gene networks for acute leukemia cell line classification. PLoS One. 2025 May 8;20(5):e0321549. doi: 10.1371/journal.pone.0321549. eCollection 2025. PMID: 40338916 [download]
- Park H, Wang QS, Hasegawa T, Namkoong H, Tanaka H, Koike R, Kitagawa Y, Kimura A, Imoto S, Kanai T, Fukunaga K, Ogawa S, Okada Y, Miyano S. Unraveling the COVID-19 Severity Hubs and Interplays in Inflammatory-Related RNA-Protein Networks. Int J Mol Sci. 2025 May 6;26(9):4412. doi: 10.3390/ijms26094412. PMID: 40362649 [download]
- Sasa N, Kojima S, Koide R, Hasegawa T, Namkoong H, Hirota T, Watanabe R, Nakamura Y, Oguro-Igashira E, Ogawa K, Yata T, Sonehara K, Yamamoto K, Kishikawa T, Sakaue S, Edahiro R, Shirai Y, Maeda Y, Nii T, Chubachi S, Tanaka H, Yabukami H, Suzuki A, Nakajima K, Arase N, Okamoto T, Nishikawa R, Namba S, Naito T, Miyagawa I, Tanaka H, Ueno M, Ishitsuka Y, Furuta J, Kunimoto K, Kajihara I, Fukushima S, Miyachi H, Matsue H, Kamata M, Momose M, Bito T, Nagai H, Ikeda T, Horikawa T, Adachi A, Matsubara T, Ikumi K, Nishida E, Nakagawa I, Yagita-Sakamaki M, Yoshimura M, Ohshima S, Kinoshita M, Ito S, Arai T, Hirose M, Tanino Y, Nikaido T, Ichiwata T, Ohkouchi S, Hirano T, Takada T, Tazawa R, Morimoto K, Takaki M, Konno S, Suzuki M, Tomii K, Nakagawa A, Handa T, Tanizawa K, Ishii H, Ishida M, Kato T, Takeda N, Yokomura K, Matsui T, Uchida A, Inoue H, Imaizumi K, Goto Y, Kida H, Fujisawa T, Suda T, Yamada T, Satake Y, Ibata H, Saigusa M, Shirai T, Hizawa N, Nakata K; Japan COVID-19 Task Force; Imafuku S, Tada Y, Asano Y, Sato S, Nishigori C, Jinnin M, Ihn H, Asahina A, Saeki H, Kawamura T, Shimada S, Katayama I, Poisner HM, Mack TM, Bick AG, Higasa K, Okuno T, Mochizuki H, Ishii M, Koike R, Kimura A, Noguchi E, Sano S, Inohara H, Fujimoto M, Inoue Y, Yamaguchi E, Ogawa S, Kanai T, Morita A, Matsuda F, Tamari M, Kumanogoh A, Tanaka Y, Ohmura K, Fukunaga K, Imoto S, Miyano S, Parrish NF, Okada Y. Nat Genet. 2025 Jan;57(1):65-79. doi: 10.1038/s41588-024-02022-z. Epub 2025 Jan 3. PMID: 3975377 [download]
- Shimada T, Maetani T, Chubachi S, Tanabe N, Asakura T, Namkoong H, Tanaka H, Azekawa S, Otake S, Nakagawara K, Fukushima T, Watase M, Shiraishi Y, Terai H, Sasaki M, Ueda S, Kato Y, Harada N, Suzuki S, Yoshida S, Tateno H, Shimizu K, Sato S, Yamada Y, Jinzaki M, Hirai T, Okada Y, Koike R, Ishii M, Kimura A, Imoto S, Miyano S, Ogawa S, Kanai T, Fukunaga K. Erector Spinae Muscle to Epicardial Visceral Fat Ratio on Chest CT Predicts the Severity of Coronavirus Disease 2019. J Cachexia Sarcopenia Muscle. 2025 Feb;16(1):e13721. doi: 10.1002/jcsm.13721. PMID: 39868664 [download]
- Sonehara K, Uwamino Y, Saiki R, Takeshita M, Namba S, Uno S, Nakanishi T, Nishimura T, Naito T, Sato G, Kanai M, Liu A, Uchida S, Kurafuji T, Tanabe A, Arai T, Ohno A, Shibata A, Tanaka S, Wakui M, Kashimura S, Tomi C, Hara A, Yoshikawa S, Gotanda K, Misawa K, Tanaka H, Azekawa S, Wang QS, Edahiro R, Shirai Y, Yamamoto K, Nagao G, Suzuki T, Kiyoshi M, Ishii-Watabe A, Higashiue S, Kobayashi S, Yamaguchi H, Okazaki Y, Matsumoto N, Masumoto A, Koga H, Kanai A; Japan COVID-19 Task Force; Biobank Japan Project; Oda Y, Suzuki Y, Matsuda K, Kitagawa Y, Koike R, Kimura A, Kumanogoh A, Yoshimura A, Imoto S, Miyano S, Kanai T, Fukunaga K, Hasegawa N, Murata M, Matsushita H, Ogawa S, Okada Y, Namkoong H. Germline variants and mosaic chromosomal alterations affect COVID-19 vaccine immunogenicity. Cell Genom. 2025 Mar 12;5(3):100783. doi: 10.1016/j.xgen.2025.100783. Epub 2025 Mar 4. PMID: 40043710 [download]
- Suehara Y, Sakamoto K, Fujisawa M, Fukumoto K, Abe Y, Makishima K, Suma S, Sakamoto T, Hattori K, Sugio T, Kato K, Akashi K, Matsue K, Narita K, Takeuchi K, Carreras J, Nakamura N, Chiba K, Shiraishi Y, Miyano S, Ogawa S, Chiba S, Sakata-Yanagimoto M. Discrete genetic subtypes and tumor microenvironment signatures correlate with peripheral T-cell lymphoma outcomes. Leukemia. 2025 May;39(5):1184-1195. doi: 10.1038/s41375-025-02563-0. Epub 2025 Mar 31. PMID: 40164718 [download]
- Takahashi Y, Wang QS, Hasegawa T, Namkoong H, Inoue F, Fukunaga K, Imoto S, Miyano S, Okada Y; Japan COVID-19 Task Force. JOB: Japan Omics Browser provides integrative visualization of multi-omics data. BMC Genomics. 2025 May 7;26(1):451. doi: 10.1186/s12864-025-11639-1. PMID: 40335902 [download]
- Watase M, Shiraishi Y, Chubachi S, Tanabe N, Maetani T, Asakura T, Namkoong H, Tanaka H, Shimada T, Azekawa S, Otake S, Fukushima T, Nakagawara K, Masaki K, Terai H, Mochimaru T, Sasaki M, Ueda S, Kato Y, Harada N, Suzuki S, Yoshida S, Tateno H, Yamada Y, Jinzaki M, Okada Y, Koike R, Ishii M, Kimura A, Imoto S, Miyano S, Ogawa S, Kanai T, Fukunaga K. Coronary Artery Calcification on Chest Computed Tomography as a Predictor of Cardiovascular Adverse Events in Patients With COVID-19 - A Multicenter Retrospective Study in Japan. Circ J. 2025 Feb 25;89(3):373-381. doi: 10.1253/circj.CJ-24-0661. Epub 2025 Jan 18. PMID: 39828330 [download]
- Yata T, Sato G, Ogawa K, Naito T, Sonehara K, Saiki R, Edahiro R, Namba S, Watanabe M, Shirai Y, Yamamoto K, NamKoong H, Nakanishi T, Yamamoto Y, Hosokawa A, Yamamoto M; Japan MS/NMOSD biobank; BioBank Japan Project; Japan COVID-19 Task Force; Oguro-Igashira E, Nii T, Maeda Y, Nakajima K, Nishikawa R, Tanaka H, Nakayamada S, Matsuda K, Nishigori C, Sano S, Kinoshita M, Koike R, Kimura A, Imoto S, Miyano S, Fukunaga K, Mihara M, Shimizu Y, Kawachi I, Miyamoto K, Tanaka Y, Kumanogoh A, Niino M, Nakatsuji Y, Ogawa S, Matsushita T, Kira JI, Mochizuki H, Isobe N, Okuno T, Okada Y. Contribution of germline and somatic mutations to risk of neuromyelitis optica spectrum disorder. Cell Genom. 2025 Mar 12;5(3):100776. doi: 10.1016/j.xgen.2025.100776. Epub 2025 Feb 21. PMID: 39986280 [download]
- Ashida S, Kawada C, Tanaka H, Kurabayashi A, Yagyu KI, Sakamoto S, Maejima K, Miyano S, Daibata M, Nakagawa H, Inoue K. Cutibacterium acnes invades prostate epithelial cells to induce BRCAness as a possible pathogen of prostate cancer. Prostate. 2024 Aug;84(11):1056-1066. doi: 10.1002/pros.24723. PMID: 38721925 Epub 2024 May 9. [download]
- Azekawa S, Maetani T, Chubachi S, Asakura T, Tanabe N, Shiraishi Y, Namkoong H, Tanaka H, Shimada T, Fukushima T, Otake S, Nakagawara K, Watase M, Terai H, Sasaki M, Ueda S, Kato Y, Harada N, Suzuki S, Yoshida S, Tateno H, Yamada Y, Jinzaki M, Hirai T, Okada Y, Koike R, Ishii M, Kimura A, Imoto S, Miyano S, Ogawa S, Kanai T, Fukunaga K. CT-derived vertebral bone mineral density is a useful biomarker to predict COVID-19 outcome. Bone. 2024 Apr 8;184:117095. doi: 10.1016/j.bone.2024.117095. PMID: 38599262 [download]
- Fukushima T, Maetani T, Chubachi S, Tanabe N, Asakura T, Namkoong H, Tanaka H, Shimada T, Azekawa S, Otake S, Nakagawara K, Watase M, Shiraishi Y, Terai H, Sasaki M, Ueda S, Kato Y, Harada N, Suzuki S, Yoshida S, Tateno H, Yamada Y, Jinzaki M, Hirai T, Okada Y, Koike R, Ishii M, Kimura A, Imoto S, Miyano S, Ogawa S, Kanai T, Fukunaga K. Epicardial adipose tissue measured from analysis of adipose tissue area using chest CT imaging is the best potential predictor of COVID-19 severity. Metabolism. 2024 Jan;150:155715. doi: 10.1016/j.metabol.2023.155715. Epub 2023 Oct 31. PMID: 37918794 [download]
- Ito S, Ono K, Miyano S. Large-scale WGS Analysis on the supercomputer Fugaku. Proceedings of the 2024 14th International Conference on Bioscience, Biochemistry and Bioinformatics (ICBBB 2024). 2024 Jan; 14-20. doi: https://doi.org/10.1145/3640900.3640903. [download]
- Iyoda S, Yoshida K, Shoji K, Ito N, Tanaka M, Nannya Y, Yamato G, Tsujimoto S, Shiba N, Hayashi Y, Shiozawa Y, Shiraishi Y, Chiba K, Okada A, Tanaka H, Miyano S, Koga Y, Goto H, Moritake H, Terui K, Ito E, Kiyokawa N, Tomizawa D, Taga T, Tawa A, Takita J, Nishikori M, Adachi S, Ogawa S, Matsuo H. KRAS G12 mutations as adverse prognostic factors in KMT2A-rearranged acute myeloid leukemia. Leukemia. 2024 Jul;38(7):1609-1612. Epub 2024 Apr 18. doi: 10.1038/s41375-024-02244-4. PMID: 38632314 [download]
- Maeda-Minami A, Yoshino T, Katayama K, Horiba Y, Hikiami H, Shimada Y, Namiki T, Tahara E, Minamizawa K, Muramatsu SI, Yamaguchi R, Imoto S, Miyano S, Mima H, Uneda K, Nogami T, Fukunaga K, Watanabe K. Machine learning model for predicting the cold-heat pattern in Kampo medicine: a multicenter prospective observational study. Front Pharmacol. 2024 Oct 25;15:1412593. doi: 10.3389/fphar.2024.1412593. eCollection 2024. PMID: 39525633 [download]
- Maruhashi K, Kashima H, Miyano S, Park H. Meta graphical lasso: uncovering hidden interactions among latent mechanisms. Sci Rep. 2024 Aug 5;14(1):18105. doi: 10.1038/s41598-024-68959-7. PMID: 39103384 [download]
- Matsubara Y, Kiyohara H, Mikami Y, Nanki K, Namkoong H, Chubachi S, Tanaka H, Azekawa S, Sugimoto S, Yoshimatsu Y, Sujino T, Takabayashi K, Hosoe N, Sato T, Ishii M, Hasegawa N, Okada Y, Koike R, Kitagawa Y, Kimura A, Imoto S, Miyano S, Ogawa S, Fukunaga K, Kanai T; Japan COVID-19 Task Force. Gastrointestinal symptoms in COVID-19 and disease severity: a Japanese registry-based retrospective cohort study. J Gastroenterol. 2024 Mar;59(3):195-208. doi: 10.1007/s00535-023-02071-x. Epub 2024 Jan 25. PMID: 38270615 [download]
- Nakagawara K, Shiraishi Y, Chubachi S, Tanabe N, Maetani T, Asakura T, Namkoong H, Tanaka H, Shimada T, Azekawa S, Otake S, Fukushima T, Watase M, Terai H, Sasaki M, Ueda S, Kato Y, Harada N, Suzuki S, Yoshida S, Tateno H, Yamada Y, Jinzaki M, Hirai T, Okada Y, Koike R, Ishii M, Kimura A, Imoto S, Miyano S, Ogawa S, Kanai T, Fukunaga K. Integrated assessment of computed tomography density in pectoralis and erector spinae muscles as a prognostic biomarker for coronavirus disease 2019. Clin Nutr. 2024 Mar;43(3):815-824. doi: 10.1016/j.clnu.2024.02.004. PMID: 38350289 [download]
- Okawa Y, Sasagawa S, Kato H, Johnson TA, Nagaoka K, Kobayashi Y, Hayashi A, Shibayama T, Maejima K, Tanaka H, Miyano S, Shibahara J, Nishizuka S, Hirano S, Seto Y, Iwaya T, Kakimi K, Yasuda T, Nakagawa H. Immuno-genomic analysis reveals eosinophilic feature and favorable prognosis of female non-smoking esophageal squamous cell carcinomas. Cancer Lett. 2024 Jan 28;581:216499. Epub 2023 Nov 25. PMID: 38013050 doi: 10.1016/j.canlet.2023.216499. [download]
- Otake S, Shiraishi Y, Chubachi S, Tanabe N, Maetani T, Asakura T, Namkoong H, Shimada T, Azekawa S, Nakagawara K, Tanaka H, Fukushima T, Watase M, Terai H, Sasaki M, Ueda S, Kato Y, Harada N, Suzuki S, Yoshida S, Tateno H, Yamada Y, Jinzaki M, Hirai T, Okada Y, Koike R, Ishii M, Hasegawa N, Kimura A, Imoto S, Miyano S, Ogawa S, Kanai T, Fukunaga K. Lung volume measurement using chest CT in COVID-19 patients: a cohort study in Japan. BMJ Open Respir Res. 2024 Apr 24;11(1):e002234. doi: 10.1136/bmjresp-2023-002234. PMID: 38663888 [download]
- Park H, Miyano S. Differential Gene Regulatory Network Analysis between Azacitidine-Sensitive and -Resistant Cell Lines. Int J Mol Sci. 2024 Mar 14;25(6):3302. doi: 10.3390/ijms25063302. PMID: 38542276 [download]
- Park H, Miyano S. Network-Constrained Eigen-Single-Cell Profile Estimation for Uncovering Crucial Immunogene Regulatory Systems in Human Bone Marrow. J Comput Biol. 2024 Nov;31(11):1158-1178. Epub 2024 Sep 6. doi: 10.1089/cmb.2024.0539. PMID: 39239711 [download]
- Park H, Miyano S. Sparse spectral graph analysis and its application to gastric cancer drug resistance-specific molecular interplays identification. PLoS One. 2024 Jul 5;19(7):e0305386. doi: 10.1371/journal.pone.0305386. eCollection 2024. PMID: 38968283 doi: 10.1371/journal.pone.0305386 [download]
- Pérez-Saldívar M, Nakamura Y, Kiyotani K, Imoto S, Katayama K, Yamaguchi R, Miyano S, Martínez-Barnetche J, Godoy-Lozano EE, Ordoñez G, Sotelo J, González-Conchillos H, Martínez-Palomo A, Flores-Rivera J, Santos-Argumedo L, Sánchez-Salguero ES, Espinosa-Cantellano M. Comparative analysis of the B cell receptor repertoire during relapse and remission in patients with multiple sclerosis. Clin Immunol. 2024 Nov 17;269:110398. doi: 10.1016/j.clim.2024.110398. Online ahead of print. PMID: 39551364 [download]
- Sakurai K, Chubachi S, Asakura T, Namkoong H, Tanaka H, Azekawa S, Shimada T, Otake S, Nakagawara K, Fukushima T, Lee H, Watase M, Kusumoto T, Masaki K, Kamata H, Ishii M, Hasegawa N, Okada Y, Koike R, Kitagawa Y, Kimura A, Imoto S, Miyano S, Ogawa S, Kanai T, Fukunaga K. Prognostic significance of hypertension history and blood pressure on admission in Japanese patients with coronavirus disease 2019: integrative analysis from the Japan COVID-19 Task Force. Hypertens Res. 2024 Mar;47(3):639-648. doi: 10.1038/s41440-023-01490-w. 2023 Nov 2. Online ahead of print. PMID: 37919428 [download]
- Sato T, Yoshida K, Toki T, Kanezaki R, Terui K, Saiki R, Ojima M, Ochi Y, Mizuno S, Yoshihara M, Uechi T, Kenmochi N, Tanaka S, Matsubayashi J, Kisai K, Kudo K, Yuzawa K, Takahashi Y, Tanaka T, Yamamoto Y, Kobayashi A, Kamio T, Sasaki S, Shiraishi Y, Chiba K, Tanaka H, Muramatsu H, Hama A, Hasegawa D, Sato A, Koh K, Karakawa S, Kobayashi M, Hara J, Taneyama Y, Imai C, Hasegawa D, Fujita N, Yoshitomi M, Iwamoto S, Yamato G, Saida S, Kiyokawa N, Deguchi T, Ito M, Matsuo H, Adachi S Prof, Hayashi Y, Taga T, Moriya Saito A, Horibe K, Watanabe K, Tomizawa D, Miyano S, Takahashi S, Ogawa S, Ito E. Landscape of driver mutations and their clinical effects on Down syndrome-related myeloid neoplasms. Blood. 2024 Jun 20;143(25):2627-2643. doi: 10.1182/blood.2023022247. PMID: 38513239 [download]
- Tanaka H, Chubachi S, Asakura T, Namkoong H, Azekawa S, Otake S, Nakagawara K, Fukushima T, Lee H, Watase M, Sakurai K, Kusumoto T, Masaki K, Kamata H, Ishii M, Hasegawa N, Okada Y, Koike R, Kitagawa Y, Kimura A, Imoto S, Miyano S, Ogawa S, Kanai T, Fukunaga K. Prognostic significance of chronic kidney disease and impaired renal function in Japanese patients with COVID-19. BMC Infect Dis. 2024 May 25;24(1):527. doi: 10.1186/s12879-024-09414-w. PMID: 38796423 [download]
- Tanaka H, Toya E, Chubachi S, Namkoong H, Asakura T, Azekawa S, Otake S, Nakagawara K, Fukushima T, Watase M, Sakurai K, Masaki K, Kamata H, Ishii M, Hasegawa N, Okada Y, Koike R, Kitagawa Y, Kimura A, Imoto S, Miyano S, Ogawa S, Kanai T, Fukunaga K. Combined use of serum ferritin and KL-6 levels as biomarkers for predicting COVID-19 severity. Respir Investig. 2024 Nov;62(6):1132-1136. doi: 10.1016/j.resinv.2024.09.011. Epub 2024 Oct 3. PMID: 39366121. [download]
- Tomofuji Y, Edahiro R, Sonehara K, Shirai Y, Kock KH, Wang QS, Namba S, Moody J, Ando Y, Suzuki A, Yata T, Ogawa K, Naito T, Namkoong H, Xuan Lin QX, Buyamin EV, Tan LM, Sonthalia R, Han KY, Tanaka H, Lee H; Asian Immune Diversity Atlas (AIDA) Network; Japan COVID-19 Task Force; Biobank Japan Project; Okuno T, Liu B, Matsuda K, Fukunaga K, Mochizuki H, Park WY, Yamamoto K, Hon CC, Shin JW, Prabhakar S, Kumanogoh A, Okada Y. Quantification of escape from X chromosome inactivation with single-cell omics data reveals heterogeneity across cell types and tissues. Cell Genom. 2024 Aug 14;4(8):100625. doi: 10.1016/j.xgen.2024.100625. Epub 2024 Jul 30. PMID: 39084228 [download]
- Wang QS, Hasegawa T, Namkoong H, Saiki R, Edahiro R, Sonehara K, Tanaka H, Azekawa S, Chubachi S, Takahashi Y, Sakaue S, Namba S, Yamamoto K, Shiraishi Y, Chiba K, Tanaka H, Makishima H, Nannya Y, Zhang Z, Tsujikawa R, Koike R, Takano T, Ishii M, Kimura A, Inoue F, Kanai T, Fukunaga K, Ogawa S, Imoto S, Miyano S, Okada Y; Japan COVID-19 Task Force. Statistically and functionally fine-mapped blood eQTLs and pQTLs from 1,405 humans reveal distinct regulation patterns and disease relevance. Nat Genet. 2024 Oct;56(10)2054-2067. doi: 10.1038/s41588-024-01896-3. Epub 2024 Sep 24. doi: 10.1038/s41588-024-01896-3. Epub ahead of print. Publisher Correction: Nat Genet. 2024 Oct;56(10):2281. Epub ahead of print. 2024 Sep 27. doi: 10.1038/s41588-024-01959-5. PMID: 39317738. [download]
- Watanabe K, Tasaka K, Ogata H, Kato S, Ueno H, Umeda K, Isobe T, Kubota Y, Sekiguchi M, Kimura S, Sato-Otsubo A, Hiwatari M, Ushiku T, Kato M, Oka A, Miyano S, Ogawa S, Takita J. Inhibition of the galactosyltransferase C1GALT1 reduces osteosarcoma cell proliferation by interfering with ERK signaling and cell cycle progression. Cancer Gene Ther. 2024 Jul;31(7):1049-1059. doi: 10.1038/s41417-024-00773-9. PMID: 38622340 [download]
- Azekawa S, Chubachi S, Asakura T, Namkoong H, Sato Y, Edahiro R, Lee H, Tanaka H, Otake S, Nakagawara K, Fukushima T, Watase M, Sakurai K, Kusumoto T, Masaki K, Kamata H, Ishii M, Hasegawa N, Okada Y, Koike R, Kitagawa Y, Kimura A, Imoto S, Miyano S, Ogawa S, Kanai T, Fukunaga K; Japan COVID-19 Task Force. Serum KL-6 levels predict clinical outcomes and are associated with MUC1 polymorphism in Japanese patients with COVID-19. BMJ Open Respir Res. 2023 May;10(1):e001625. doi: 10.1136/bmjresp-2023-001625. PMID: 37230764 [download]
- COVID-19 Host Genetics Initiative. A second update on mapping the human genetic architecture of COVID-19. Nature. 2023 Sep;621(7977):E7-E26. doi: 10.1038/s41586-023-06355-3. Epub 2023 Sep 6. PMID: 37674002 [download]
- Kobayashi Y, Niida A, Nagayama S, Saeki K, Haeno H, Takahashi KK, Hayashi S, Ozato Y, Saito H, Hasegawa T, Nakamura H, Tobo T, Kitagawa A, Sato K, Shimizu D, Hirata H, Hisamatsu Y, Toshima T, Yonemura Y, Masuda T, Mizuno S, Kawazu M, Kohsaka S, Ueno T, Mano H, Ishihara S, Uemura M, Mori M, Doki Y, Eguchi H, Oshima M, Suzuki Y, Shibata T, Mimori K. Subclonal accumulation of immune escape mechanisms in microsatellite instability-high colorectal cancers. Br J Cancer. 2023 Oct;129:1105-1118. doi: 10.1038/s41416-023-02395-8. Epub 2023 Aug 18. PMID: 37596408; PMCID: PMC10539316.
- Latorre AAE, Nakamura K, Seino K, Hasegawa T. Vector autoregression for forecasting the number of COVID-19 cases and analyzing behavioral indicators in the Philippines: ecologic time-trend study. JMIR Form Res. 2023 Jun 27;7:e46357. doi: 10.2196/46357. PMID: 37368473; PMCID: PMC10337462.
- Sera T, Otani N, Bannai H, Hasegawa T, Umemura T, Honda H, Kimura A. The current status of emergency departments in secondary emergency medical institutions in Japan: a questionnaire survey. Int J Emerg Med. 2023 Jun 23;16:40. doi: 10.1186/s12245-023-00513-0. PMID: 37353768; PMCID: PMC10290351.
- Edahiro R, Shirai Y, Takeshima Y, Sakakibara S, Yamaguchi Y, Murakami T, Morita T, Kato Y, Liu YC, Motooka D, Naito Y, Takuwa A, Sugihara F, Tanaka K, Wing JB, Sonehara K, Tomofuji Y; Japan COVID-19 Task Force; Namkoong H, Tanaka H, Lee H, Fukunaga K, Hirata H, Takeda Y, Okuzaki D, Kumanogoh A, Okada Y. Single-cell analyses and host genetics highlight the role of innate immune cells in COVID-19 severity. Nat Genet. 2023 May;55(5):753-767. doi: 10.1038/s41588-023-01375-1. Epub 2023 Apr 24. PMID: 37095364. [download]
- Fukushima T, Chubachi S, Namkoong H, Asakura T, Tanaka H, Lee H, Azekawa S, Okada Y, Koike R, Kimura A, Imoto S, Miyano S, Ogawa S, Kanai T, Fukunaga K; Japan COVID-19 Task Force. Clinical significance of prediabetes, undiagnosed diabetes and diagnosed diabetes on critical outcomes in COVID-19: Integrative analysis from the Japan COVID-19 task force. Diabetes Obes Metab. 2023 Jan; 25(1):144-155. doi: 10.1111/dom.14857. PMID: 36056760. https://dom-pubs.onlinelibrary.wiley.com/doi/10.1111/dom.14857 [download]
- Hara Y, Shiba N, Yoshida K, Yamato G, Kaburagi T, Shiraishi Y, Ohki K, Shiozawa Y, Kawamura M, Kawasaki H, Sotomatsu M, Takizawa T, Matsuo H, Shimada A, Kiyokawa N, Tomizawa D, Taga T, Ito E, Horibe K, Miyano S, Adachi S, Taki T, Ogawa S, Hayashi Y. TP53 and RB1 alterations characterize poor prognostic subgroups in pediatric acute myeloid leukemia. Genes Chromosomes Cancer. 2023 Jul;62(7):412-422. doi: 10.1002/gcc.23147. Epub 2023 Apr 27. PMID: 3710230 [download]
- Ito S, Miyano S, Ono K. Acceleration of BAM I/O on distributed file systems. Proc. 2023 IEEE International Conference on Bioinformatics and Biomedicine (BIBM). 2023 Dec; 3299-3304. https://doi.org/10.1109/bibm58861.2023.10385370 https://ieeexplore.ieee.org/document/10385370
- Kaburagi T, Shiba N, Yamato G, Yoshida K, Tabuchi K, Ohki K, Ishikita E, Hara Y, Shiraishi Y, Kawasaki H, Sotomatsu M, Takizawa T, Taki T, Kiyokawa N, Tomizawa D, Horibe K, Miyano S, Taga T, Adachi S, Ogawa S, Hayashi Y. UBTF-Internal tandem duplication as a novel poor prognostic factor in pediatric acute myeloid leukemia. Genes Chromosomes Cancer. 2023 Apr;62(4):202-209. doi: 10.1002/gcc.23110. PMID: 36448876. https://onlinelibrary.wiley.com/doi/10.1002/gcc.23110 [download]
- Kawachi K, Tang X, Kasajima R, Yamanaka T, Shimizu E, Katayama K, Yamaguchi R, Yokoyama K, Yamaguchi K, Furukawa Y, Miyano S, Imoto S, Yoshioka E, Washimi K, Okubo Y, Sato S, Yokose T, Miyagi Y. Genetic analysis of low-grade adenosquamous carcinoma of the breast progressing to high-grade metaplastic carcinoma. Breast Cancer Res Treat. 2023 Dec;202(3):563-573. 2023 Aug 31. doi: 10.1007/s10549-023-07078-9. Online ahead of print. PMID: 37650999 [download]
- Kitagawa A, Osawa T, Noda M, Kobayashi Y, Aki S, Nakano Y, Saito T, Shimizu D, Komatsu H, Sugaya M, Takahashi J, Kosai K, Takao S, Motomura Y, Sato K, Hu Q, Fujii A, Wakiyama H, Tobo T, Uchida H, Sugimachi K, Shibata K, Utsunomiya T, Kobayashi S, Ishii H, Hasegawa T, Masuda T, Matsui Y, Niida A, Soga T, Suzuki Y, Miyano S, Aburatani H, Doki Y, Eguchi H, Mori M, Nakayama KI, Shimamura T, Shibata T, Mimori K. Convergent genomic diversity and novel BCAA metabolism in intrahepatic cholangiocarcinoma. Br J Cancer. 2023 Jun;128(12):2206-2217. doi: 10.1038/s41416-023-02256-4. Epub 2023 Apr 19. PMID: 37076565 [download]
- Kusumoto T, Chubachi S, Namkoong H, Tanaka H, Lee H, Azekawa S, Otake S, Nakagawara K, Fukushima T, Morita A, Watase M, Sakurai K, Asakura T, Masaki K, Kamata H, Ishii M, Hasegawa N, Harada N, Ueda T, Ueda S, Ishiguro T, Arimura K, Saito F, Yoshiyama T, Nakano Y, Mutoh Y, Suzuki Y, Edahiro R, Sano H, Sato Y, Okada Y, Koike R, Kitagawa Y, Tokunaga K, Kimura A, Imoto S, Miyano S, Ogawa S, Kanai T, Fukunaga K. Association between ABO blood group/genotype and COVID-19 in a Japanese population. Ann Hematol. 2023 Nov;102(11):3239-3249. Epub 2023 Aug 15. doi: 10.1007/s00277-023-05407-y. PMID: 37581712 [download]
- Kusumoto T, Chubachi S, Namkoong H, Tanaka H, Lee H, Otake S, Nakagawara K, Fukushima T, Morita A, Watase M, Asakura T, Masaki K, Kamata H, Ishii M, Hasegawa N, Harada N, Ueda T, Ueda S, Ishiguro T, Arimura K, Saito F, Yoshiyama T, Nakano Y, Mutoh Y, Suzuki Y, Edahiro R, Murakami K, Sato Y, Okada Y, Koike R, Kitagawa Y, Tokunaga K, Kimura A, Imoto S, Miyano S, Ogawa S, Kanai T, Fukunaga K. Characteristics of patients with COVID-19 who have deteriorating chest X-ray findings within 48 h: a retrospective cohort study. Sci Rep. 2023 Dec 12;13(1):22054. doi: 10.1038/s41598-023-49340-6. PMID: 38086863 [download]
- Makishima H, Saiki R, Nannya Y, Korotev SC, Gurnari C, Takeda J, Momozawa Y, Best S, Krishnamurthy P, Yoshizato T, Atsuta Y, Shiozawa Y, Iijima-Yamashita Y, Yoshida K, Shiraishi Y, Nagata Y, Kakiuchi N, Onizuka M, Chiba K, Tanaka H, Kon A, Ochi Y, Nakagawa MM, Okuda R, Mori T, Yoda A, Itonaga H, Miyazaki Y, Sanada M, Ishikawa T, Chiba S, Tsurumi H, Kasahara S, Müller-Tidow C, Takaori-Kondo A, Ohyashiki K, Kiguchi T, Matsuda F, Jansen JH, Polprasert C, Blombery P, Kamatani Y, Miyano S, Malcovati L, Haferlach T, Kubo M, Cazzola M, Kulasekararaj AG, Godley LA, Maciejewski JP, Ogawa S. Germline d mutations define a unique subtype of myeloid neoplasms. Blood. 2023 Feb 2;141(5):534-549. doi: 10.1182/blood.2022018221. PMID: 36322930. [download]
- Nakagawara K, Kamata H, Chubachi S, Namkoong H, Tanaka H, Lee H, Otake S, Fukushima T, Kusumoto T, Morita A, Azekawa S, Watase M, Asakura T, Masaki K, Ishii M, Endo A, Koike R, Ishikura H, Takata T, Matsushita Y, Harada N, Kokutou H, Yoshiyama T, Kataoka K, Mutoh Y, Miyawaki M, Ueda S, Ono H, Ono T, Shoko T, Muranaka H, Kawamura K, Mori N, Mochimaru T, Fukui M, Chihara Y, Nagasaki Y, Okamoto M, Amishima M, Odani T, Tani M, Nishi K, Shirai Y, Edahiro R, Ando A, Hashimoto N, Ogura S, Kitagawa Y, Kita T, Kagaya T, Kimura Y, Miyazawa N, Tsuchida T, Fujitani S, Murakami K, Sano H, Sato Y, Tanino Y, Otsuki R, Mashimo S, Kuramochi M, Hosoda Y, Hasegawa Y, Ueda T, Takaku Y, Ishiguro T, Fujiwara A, Kuwahara N, Kitamura H, Hagiwara E, Nakamori Y, Saito F, Kono Y, Abe S, Ishii T, Ohba T, Kusaka Y, Watanabe H, Masuda M, Watanabe H, Kimizuka Y, Kawana A, Kasamatsu Y, Hashimoto S, Okada Y, Takano T, Katayama K, Ai M, Kumanogoh A, Sato T, Tokunaga K, Imoto S, Kitagawa Y, Kimura A, Miyano S, Hasegawa N, Ogawa S, Kanai T, Fukunaga K; Japan COVID-19 Task Force. Impact of respiratory bacterial infections on mortality in Japanese patients with COVID-19: a retrospective cohort study. BMC Pulm Med. 2023 Apr 26;23(1):146. doi: 10.1186/s12890-023-02418-3. PMID: 37101265 [download]
- Nakagawara K, Kamata H, Chubachi S, Namkoong H, Tanaka H, Lee H, Otake S, Fukushima T, Kusumoto T, Morita A, Azekawa S, Watase M, Asakura T, Masaki K, Ishii M, Endo A, Koike R, Ishikura H, Takata T, Matsushita Y, Harada N, Kokutou H, Yoshiyama T, Kataoka K, Mutoh Y, Miyawaki M, Ueda S, Ono H, Ono T, Shoko T, Muranaka H, Kawamura K, Mori N, Mochimaru T, Fukui M, Chihara Y, Nagasaki Y, Okamoto M, Amishima M, Odani T, Tani M, Nishi K, Shirai Y, Edahiro R, Ando A, Hashimoto N, Ogura S, Kitagawa Y, Kita T, Kagaya T, Kimura Y, Miyazawa N, Tsuchida T, Fujitani S, Murakami K, Sano H, Sato Y, Tanino Y, Otsuki R, Mashimo S, Kuramochi M, Hosoda Y, Hasegawa Y, Ueda T, Takaku Y, Ishiguro T, Fujiwara A, Kuwahara N, Kitamura H, Hagiwara E, Nakamori Y, Saito F, Kono Y, Abe S, Ishii T, Ohba T, Kusaka Y, Watanabe H, Masuda M, Watanabe H, Kimizuka Y, Kawana A, Kasamatsu Y, Hashimoto S, Okada Y, Takano T, Katayama K, Ai M, Kumanogoh A, Sato T, Tokunaga K, Imoto S, Kitagawa Y, Kimura A, Miyano S, Hasegawa N, Ogawa S, Kanai T, Fukunaga K; Japan COVID-19 Task Force. Diagnostic significance of secondary bacteremia in patients with COVID-19. J Infect Chemother. 2023 Apr;29(4):422-426. doi: 10.1016/j.jiac.2023.01.006. Epub 2023 Jan 20. PMID: 36682606 [download]
- Nannya Y, Tobiasson M, Sato S, Bernard E, Ohtake S, Takeda J, Creignou M, Zhao L, Kusakabe M, Shibata Y, Nakamura N, Watanabe M, Hiramoto N, Shiozawa Y, Shiraishi Y, Tanaka H, Yoshida K, Kakiuchi N, Makishima H, Nakagawa MM, Usuki K, Watanabe M, Imada K, Handa H, Taguchi M, Kiguchi T, Ohyashiki K, Ishikawa T, Takaori-Kondo A, Tsurumi H, Kasahara S, Chiba S, Naoe T, Miyano S, Papaemmanuil E, Miyazaki Y, Hellström Lindberg E, Ogawa S. Post-azacitidine clone size predicts outcome of patients with myelodysplastic syndromes and related myeloid neoplasms. Blood Adv. 2023 Jul 25;7(14):3624-3636. doi: 10.1182/bloodadvances.2022009564. Online ahead of print. PMID: 36989067 [download]
- Nishimura T, Kakiuchi N, Yoshida K, Sakurai T, Kataoka TR, Kondoh E, Chigusa Y, Kawai M, Sawada M, Inoue T, Takeuchi Y, Maeda H, Baba S, Shiozawa Y, Saiki R, Nakagawa MM, Nannya Y, Ochi Y, Hirano T, Nakagawa T, Inagaki-Kawata Y, Aoki K, Hirata M, Nanki K, Matano M, Saito M, Suzuki E, Takada M, Kawashima M, Kawaguchi K, Chiba K, Shiraishi Y, Takita J, Miyano S, Mandai M, Sato T, Takeuchi K, Haga H, Toi M, Ogawa S. Evolutionary histories of breast cancer and related clones. Nature. 2023 Aug;620(7974):607-614. doi: 10.1038/s41586-023-06333-9. Epub 2023 Jul 26. PMID: 37495687 [download]
- Ozawa T, Asakura T, Chubachi S, Namkoong H, Tanaka H, Lee K, Fukushima T, Otake S, Nakagawara K, Watase M, Masaki K, Kamata H, Ishii M, Hasegawa N, Harada N, Ueda T, Ueda S, Ishiguro T, Arimura K, Saito F, Yoshiyama T, Nakano Y, Mutoh Y, Suzuki Y, Edahiro R, Murakami K, Okada Y, Koike R, Kitagawa Y, Tokunaga K, Kimura A, Imoto S, Miyano S, Ogawa S, Kanai T, Fukunaga K; Japan COVID-19 Task Force. Use of the neutrophil-to-lymphocyte ratio and an oxygen requirement to predict disease severity in patients with COVID-19. Respir Investig. 2023 Apr 19;61(4):454-459. doi: 10.1016/j.resinv.2023.03.007. Online ahead of print. PMID: 37121116 [download]
- Park H, Imoto S, Miyano S. Comprehensive information-based differential gene regulatory networks analysis (CIdrgn): Application to gastric cancer and chemotherapy-responsive gene network identification. PLoS One. 2023 Aug 23;18(8):e0286044. doi: 10.1371/journal.pone.0286044. eCollection 2023. PMID: 37610997 [download]
- Park H, Imoto S, Miyano S. Gene Regulatory Network-Classifier: Gene Regulatory Network-Based Classifier and Its Applications to Gastric Cancer Drug (5-Fluorouracil) Marker Identification. J Comput Biol. 2023 Feb;30(2):223-243. doi: 10.1089/cmb.2022.0181. [download]
- Suzuki M, Kasajima R, Yokose T, Shimizu E, Hatakeyama S, Yamaguchi K, Yokoyama K, Katayama K, Yamaguchi R, Furukawa Y, Miyano S, Imoto S, Shinozaki-Ushiku A, Ushiku T, Miyagi Y. KMT2C expression and DNA homologous recombination repair factors in lung cancers with a high-grade fetal adenocarcinoma component. Transl Lung Cancer Res. 2023 Aug 30;12(8):1738-1751. doi: 10.21037/tlcr-23-137. PMID: 37691868 [download]
- Takagi M, Hoshino A, Bousset K, Röddecke J, Martin HL, Folcut I, Tomomasa D, Yang X, Kobayashi J, Sakata N, Yoshida K, Miyano S, Ogawa S, Kojima S, Morio T, Dörk T, Kanegane H. Bone Marrow Failure and Immunodeficiency Associated with Human RAD50 Variants. J Clin Immunol. 2023 Nov;43(8):2136-2145. doi: 10.1007/s10875-023-01591-8. Epub 2023 Oct 5. Online ahead of print. PMID: 37794136 [download]
- Tanaka H, Chubachi S, Asakura T, Namkoong H, Azekawa S, Otake S, Nakagawara K, Fukushima T, Lee H, Watase M, Sakurai K, Kusumoto T, Masaki K, Kamata H, Ishii M, Hasegawa N, Okada Y, Koike R, Kitagawa Y, Kimura A, Imoto S, Miyano S, Ogawa S, Kanai T, Fukunaga K. Characteristics and clinical effectiveness of COVID-19 vaccination in hospitalized patients in Omicron-dominated epidemic wave - a nationwide study in Japan. Int J Infect Dis. 2023 Jul;132: 84-88. doi: 10.1016/j.ijid.2023.04.399. Epub 2023 Apr 21. PMID: 37086866 [download]
- Tanaka H, Chubachi S, Namkoong H, Sato Y, Asakura T, Lee H, Azekawa S, Otake S, Nakagawara K, Fukushima T, Watase M, Sakurai K, Kusumoto T, Kondo Y, Masaki K, Kamata H, Ishii M, Kaneko Y, Hasegawa N, Ueda S, Sasaki M, Izumo T, Inomata M, Miyazawa N, Kimura Y, Suzuki Y, Harada N, Ichikawa M, Takata T, Ishikura H, Yoshiyama T, Kokuto H, Murakami K, Sano H, Ueda T, Kuwahara N, Fujiwara A, Ogura T, Inoue T, Asami T, Mutoh Y, Nakachi I, Baba R, Nishi K, Tani M, Kagyo J, Hashiguchi M, Oguma T, Asano K, Nishikawa M, Watanabe H, Okada Y, Koike R, Kitagawa Y, Kimura A, Imoto S, Miyano S, Ogawa S, Kanai T, Fukunaga K. Propensity-Score Matched Analysis of the Effectiveness of Baricitinib in Patients With Coronavirus Disease 2019 (COVID-19) Using Nationwide Real-World Data: An Observational Matched Cohort Study From the Japan COVID-19 Task Force. Open Forum Infect Dis. 2023 Jun 8;10(7):ofad311. doi: 10.1093/ofid/ofad311. eCollection 2023 Jul. PMID: 37441355 [download]
- Tanaka H, Maetani T, Chubachi S, Tanabe N, Shiraishi Y, Asakura T, Namkoong H, Shimada T, Azekawa S, Otake S, Nakagawara K, Fukushima T, Watase M, Terai H, Sasaki M, Ueda S, Kato Y, Harada N, Suzuki S, Yoshida S, Tateno H, Yamada Y, Jinzaki M, Hirai T, Okada Y, Koike R, Ishii M, Hasegawa N, Kimura A, Imoto S, Miyano S, Ogawa S, Kanai T, Fukunaga K. Clinical utilization of artificial intelligence-based COVID-19 pneumonia quantification using chest computed tomography - a multicenter retrospective cohort study in Japan. Respir Res. 2023 Oct 5;24(241): doi: 10.1186/s12931-023-02530-2. PMID: 37798709 [download]
- Tanaka H, Namkoong H, Chubachi S, Irie S, Uwamino Y, Lee H, Azekawa S, Otake S, Nakagawara K, Fukushima T, Watase M, Kusumoto T, Masaki K, Kamata H, Ishii M, Okada Y, Takano T, Imoto S, Koike R, Kimura A, Miyano S, Ogawa S, Kanai T, Sato TA, Fukunaga K; Japan COVID-19 Task Force. Clinical characteristics of patients with COVID-19 harboring detectable intracellular SARS-CoV-2 RNA in peripheral blood cells. Int J Infect Dis. 2023 Oct;135:41-44. doi: 10.1016/j.ijid.2023.07.030. Epub 2023 Aug 3. PMID: 37541421 [download]
- Wang QS, Edahiro R, Namkoong H, Hasegawa T, Shirai Y, Sonehara K; Japan COVID-19 Task Force; Kumanogoh A, Ishii M, Koike R, Kimura A, Imoto S, Miyano S, Ogawa S, Kanai T, Fukunaga K, Okada Y. Estimating gene-level false discovery probability improves eQTL statistical fine-mapping precision. NAR Genom Bioinform. 2023 Oct 30;5(4):lqad090. doi: 10.1093/nargab/lqad090. eCollection 2023 Dec. PMID: 37915762 [download]
- Washimi K, Kasajima R, Shimizu E, Sato S, Okubo Y, Yoshioka E, Narimatsu H, Hiruma T, Katayama K, Yamaguchi R, Yamaguchi K, Furukawa Y, Miyano S, Imoto S, Yokose T, Miyagi Y. Histological markers, sickle-shaped blood vessels, myxoid area, and infiltrating growth pattern help stratify the prognosis of patients with myxofibrosarcoma/undifferentiated sarcoma. Sci Rep. 2023 Apr 25;13(1):6744. doi: 10.1038/s41598-023-34026-w. PMID: 37185612 [download]
- Watase M, Masaki K, Chubachi S, Namkoong H, Tanaka H, Lee H, Fukushima T, Otake S, Nakagawara K, Kusumoto T, Asakura T, Kamata H, Ishii M, Hasegawa N, Oyamada Y, Harada N, Ueda T, Ueda S, Ishiguro T, Arimura K, Saito F, Yoshiyama T, Nakano Y, Mutoh Y, Suzuki Y, Edahiro R, Sano H, Sato Y, Okada Y, Koike R, Kitagawa Y, Tokunaga K, Kimura A, Imoto S, Miyano S, Ogawa S, Kanai T, Fukunaga K; Japan COVID-19 Task Force. Impact of accumulative smoking exposure and chronic obstructive pulmonary disease on COVID-19 outcomes: report based on findings from the Japan COVID-19 task force. Int J Infect Dis. 2023 Mar;128: 121-127. doi: 10.1016/j.ijid.2022.12.019. [download] Corrigendum: Int J Infect Dis. 2024 Dec 10;151:107322. doi: 10.1016/j.ijid.2024.107322. Online ahead of print. PMID: 39662225. Corrigendum: Int J Infect Dis. 2025 Feb;151:107322. doi: 10.1016/j.ijid.2024.107322. Epub 2024 Dec 10. PMID: 39662225 [download]
- Yamaguchi K, Nakagawa S, Saku A, Isobe Y, Yamaguchi R, Sheridan P, Takane K, Ikenoue T, Zhu C, Miura M, Okawara Y, Nagatoishi S, Kozuka-Hata H, Oyama M, Aikou S, Ahiko Y, Shida D, Tsumoto K, Miyano S, Imoto S, Furukawa Y. Bromodomain protein BRD8 regulates cell cycle progression in colorectal cancer cells through a TIP60-independent regulation of the pre-RC complex. iScience. 2023 Apr 1;26(4):106563. doi: 10.1016/j.isci.2023.106563. eCollection 2023 Apr 21. PMID: 37123243 [download]
- Bai Z, Zhang YZ, Miyano S, Yamaguchi R, Fujimoto K, Uematsu S, Imoto S. Identification of bacteriophage genome sequences with representation learning. Bioinformatics. 2022 Aug 3; 8(18):4264-4270. doi: 10.1093/bioinformatics/btac509. PMID: 35920769 [download]
- Butler-Laporte G, Povysil G, Kosmicki JA, Cirulli ET, Drivas T, Furini S, Saad C, Schmidt A, Olszewski P, Korotko U, Quinodoz M, Çelik E, Kundu K, Walter K, Jung J, Stockwell AD, Sloofman LG, Jordan DM, Thompson RC, Del Valle D, Simons N, Cheng E, Sebra R, Schadt EE, Kim-Schulze S, Gnjatic S, Merad M, Buxbaum JD, Beckmann ND, Charney AW, Przychodzen B, Chang T, Pottinger TD, Shang N, Brand F, Fava F, Mari F, Chwialkowska K, Niemira M, Pula S, Baillie JK, Stuckey A, Salas A, Bello X, Pardo-Seco J, Gómez-Carballa A, Rivero-Calle I, Martinón-Torres F, Ganna A, Karczewski KJ, Veerapen K, Bourgey M, Bourque G, Eveleigh RJ, Forgetta V, Morrison D, Langlais D, Lathrop M, Mooser V, Nakanishi T, Frithiof R, Hultström M, Lipcsey M, Marincevic-Zuniga Y, Nordlund J, Schiabor Barrett KM, Lee W, Bolze A, White S, Riffle S, Tanudjaja F, Sandoval E, Neveux I, Dabe S, Casadei N, Motameny S, Alaamery M, Massadeh S, Aljawini N, Almutairi MS, Arabi YM, Alqahtani SA, Al Harthi FS, Almutairi A, Alqubaishi F, Alotaibi S, Binowayn A, Alsolm EA, El Bardisy H, Fawzy M, Cai F, Soranzo N, Butterworth A; COVID-19 Host Genetics Initiative; DeCOI Host Genetics Group; GEN-COVID Multicenter Study (Italy); Mount Sinai Clinical Intelligence Center; GEN-COVID consortium (Spain); GenOMICC Consortium; Japan COVID-19 Task Force; Regeneron Genetics Center, Geschwind DH, Arteaga S, Stephens A, Butte MJ, Boutros PC, Yamaguchi TN, Tao S, Eng S, Sanders T, Tung PJ, Broudy ME, Pan Y, Gonzalez A, Chavan N, Johnson R, Pasaniuc B, Yaspan B, Smieszek S, Rivolta C, Bibert S, Bochud PY, Dabrowski M, Zawadzki P, Sypniewski M, Kaja E, Chariyavilaskul P, Nilaratanakul V, Hirankarn N, Shotelersuk V, Pongpanich M, Phokaew C, Chetruengchai W, Tokunaga K, Sugiyama M, Kawai Y, Hasegawa T, Naito T, Namkoong H, Edahiro R, Kimura A, Ogawa S, Kanai T, Fukunaga K, Okada Y, Imoto S, Miyano S, Mangul S, Abedalthagafi MS, Zeberg H, Grzymski JJ, Washington NL, Ossowski S, Ludwig KU, Schulte EC, Riess O, Moniuszko M, Kwasniewski M, Mbarek H, Ismail SI, Verma A, Goldstein DB, Kiryluk K, Renieri A, Ferreira MAR, Richards JB. Exome-wide association study to identify rare variants influencing COVID-19 outcomes: Results from the Host Genetics Initiative. PLoS Genet. 2022 Nov 3;18(11):e1010367. doi: 10.1371/journal.pgen.1010367. eCollection 2022 Nov. PMID: 36327219. https://journals.plos.org/plosgenetics/article?id=10.1371/journal.pgen.1010367 [download]
- Fukushima T, Chubachi S, Namkoong H, Otake S, Nakagawara K, Tanaka H, Lee H, Morita A, Watase M, Kusumoto T, Masaki K, Kamata H, Ishii M, Hasegawa N, Harada N, Ueda T, Ueda S, Ishiguro T, Arimura K, Saito F, Yoshiyama T, Nakano Y, Mutoh Y, Suzuki Y, Murakami K, Okada Y, Koike R, Kitagawa Y, Kimura A, Imoto S, Miyano S, Ogawa S, Kanai T, Fukunaga K; Japan COVID-19 Task Force. U-shaped association between abnormal serum uric acid levels and COVID-19 severity: reports from the Japan COVID-19 Task Force. Int J Infect Dis. 2022 Sep; 122:747-754. doi: 10.1016/j.ijid.2022.07.014. Epub 2022 Jul 8. PMID: 35811077 [download]
- Hiwatari M, Seki M, Matsuno R, Yoshida K, Nagasawa T, Sato-Otsubo A, Yamamoto S, Kato M, Watanabe K, Sekiguchi M, Miyano S, Ogawa S, Takita J. Novel TENM3-ALK fusion is an alternate mechanism for ALK activation in neuroblastoma. Oncogene. 2022 May; 41(20):2789-2797. doi: 10.1038/s41388-022-02301-1. Epub 2022 Apr 11. PMID: 35411036 [download]
- COVID-19 Host Genetics Initiative. A first update on mapping the human genetic architecture of COVID-19. Nature. 2022 Aug;608(7921):E1-E10. doi: 10.1038/s41586-022-04826-7. Epub 2022 Aug 3. PMID: 35922517 [download]
- Isobe T, Takagi M, Sato-Otsubo A, Nishimura A, Nagae G, Yamagishi C, Tamura M, Tanaka Y, Asada S, Takeda R, Tsuchiya A, Wang X, Yoshida K, Nannya Y, Ueno H, Akazawa R, Kato I, Mikami T, Watanabe K, Sekiguchi M, Seki M, Kimura S, Hiwatari M, Kato M, Fukuda S, Tatsuno K, Tsutsumi S, Kanai A, Inaba T, Shiozawa Y, Shiraishi Y, Chiba K, Tanaka H, Kotecha RS, Cruickshank MN, Ishikawa F, Morio T, Eguchi M, Deguchi T, Kiyokawa N, Arakawa Y, Koh K, Aoki Y, Ishihara T, Tomizawa D, Miyamura T, Ishii E, Mizutani S, Wilson NK, Göttgens B, Miyano S, Kitamura T, Goyama S, Yokoyama A, Aburatani H, Ogawa S, Takita J. Multi-omics analysis defines highly refractory RAS burdened immature subgroup of infant acute lymphoblastic leukemia. Nat Commun. 2022 Aug 30;13(1):4501. doi: 10.1038/s41467-022-32266-4. PMID: 36042201 [download]
- Hasegawa T, Kakuta M, Yamaguchi R, Sato N, Mikami T, Murashita K, Nakaji S, Itoh K, Imoto S. Impact of salivary and pancreatic amylase gene copy numbers on diabetes, obesity, and functional profiles of microbiome in a northern Japanese population. Sci Rep. 2022 May 10;12:7628. doi: 10.1038/s41598-022-11730-7. PMID: 35538098; PMCID: PMC9090785.
- Kaburagi T, Yamato G, Shiba N, Yoshida K, Hara Y, Tabuchi K, Shiraishi Y, Ohki K, Sotomatsu M, Arakawa H, Matsuo H, Shimada A, Taki T, Kiyokawa N, Tomizawa D, Horibe K, Miyano S, Taga T, Adachi S, Ogawa S, Hayashi Y. Clinical significance of RAS pathway alterations in pediatric acute myeloid leukemia. Haematologica. 2022 Mar 1; 107(3):583-592. doi: 10.3324/haematol.2020.269431. PMID: 33730843 [download]
- Kato H, Maezawa Y, Nishijima D, Iwamoto E, Takeda J, Kanamori T, Yamaga M, Mishina T, Takeda Y, Izumi S, Hino Y, Nishi H, Ishiko J, Takeuchi M, Kaneko H, Koshizaka M, Mimura N, Kuzuya M, Sakaida E, Takemoto M, Shiraishi Y, Miyano S, Ogawa S, Iwama A, Sanada M, Yokote K. A high prevalence of myeloid malignancies in progeria with Werner syndrome is associated with p53 insufficiency. Exp Hematol. 2022 May;109: 11-17. doi: 10.1016/j.exphem.2022.02.005. Epub 2022 Feb 28. PMID: 35240258 [download]
- Kazama S, Yokoyama K, Ueki T, Kazumoto H, Satomi H, Sumi M, Ito I, Yusa N, Kasajima R, Shimizu E, Yamaguchi R, Imoto S, Miyano S, Tanaka Y, Denda T, Ota Y, Tojo A, Kobayashi H. Case report: Common clonal origin of concurrent langerhans cell histiocytosis and acute myeloid leukemia. Front Oncol. 2022 Sep 16;12: 974307. doi: 10.3389/fonc.2022.974307. eCollection 2022. PMID: 36185232 [download]
- Kobayashi A, Ohtaka R, Toki T, Hara J, Muramatsu H, Kanezaki R, Takahashi Y, Sato T, Kamio T, Kudo K, Sasaki S, Yoshida T, Utsugisawa T, Kanno H, Yoshida K, Nannya Y, Takahashi Y, Kojima S, Miyano S, Ogawa S, Terui K, Ito E. Dyserythropoietic anaemia with an intronic GATA1 splicing mutation in patients suspected to have Diamond-Blackfan anaemia. EJHaem. 2022 Jan 10;3(1):163-167. doi: 10.1002/jha2.374. [download]
- Kogure Y, Kameda T, Koya J, Yoshimitsu M, Nosaka K, Yasunaga JI, Imaizumi Y, Watanabe M, Saito Y, Ito Y, McClure MB, Tabata M, Shingaki S, Yoshifuji K, Chiba K, Okada A, Kakiuchi N, Nannya Y, Kamiunten A, Tahira Y, Akizuki K, Sekine M, Shide K, Hidaka T, Kubuki Y, Kitanaka A, Hidaka M, Nakano N, Utsunomiya A, Sica RA, Acuna-Villaorduna A, Janakiram M, Shah UA, Ramos JC, Shibata T, Takeuchi K, Takaori-Kondo A, Miyazaki Y, Matsuoka M, Ishitsuka K, Shiraishi Y, Miyano S, Ogawa S, Ye BH, Shimoda K, Kataoka K. Whole-genome landscape of adult T-cell leukemia/lymphoma. Blood. 2022 Feb 17;139(7):967-982. doi: 10.1182/blood.2021013568. Online ahead of print. PMID: 34695199 [download]
- Lee H, Chubachi S, Namkoong H, Asakura T, Tanaka H, Otake S, Nakagawara K, Morita A, Fukushima T, Watase M, Kusumoto T, Masaki K, Kamata H, Ishii M, Hasegawa N, Harada N, Ueda T, Ueda S, Ishiguro T, Arimura K, Saito F, Yoshiyama T, Nakano Y, Mutoh Y, Suzuki Y, Murakami K, Okada Y, Koike R, Kitagawa Y, Kimura A, Imoto S, Miyano S, Ogawa S, Kanai T, Fukunaga K; Japan COVID-19 Task Force. Characteristics of hospitalized patients with COVID-19 during the first to fifth waves of infection: a report from the Japan COVID-19 Task Force. BMC Infect Dis. 2022 Dec 12;22(1):935. doi: 10.1186/s12879-022-07927-w. PMID: 36510172. [download]
- Lee H, Chubachi S, Namkoong H, Tanaka H, Otake S, Nakagawara K, Morita A, Fukushima T, Watase M, Kusumoto T, Masaki K, Kamata H, Ishii M, Hasegawa N, Harada N, Ueda T, Ueda S, Ishiguro T, Arimura K, Saito F, Yoshiyama T, Nakano Y, Mutoh Y, Suzuki Y, Murakami K, Okada Y, Koike R, Kitagawa Y, Kimura A, Imoto S, Miyano S, Ogawa S, Kanai T, Fukunaga K; Japan COVID-19 Task Force. Effects of mild obesity on outcomes in Japanese patients with COVID-19: a nationwide consortium to investigate COVID-19 host genetics. Nutr Diabetes. 2022 Aug 9;12(1):38. doi: 10.1038/s41387-022-00217-z. PMID: 35945221 [download]
- Lu L, Qin J, Chen J, Wu H, Zhao Q, Miyano S, Zhang Y, Yu H, Li C. DDIT: An Online Predictor for Multiple Clinical Phenotypic Drug-Disease Associations. Front Pharmacol. 2022 Jan 19;12: 772026. doi: 10.3389/fphar.2021.772026. PMID: 35126114 [download]
- Lu L, Qin J, Chen J, Yu N, Miyano S, Deng Z, Li C. Recent computational drug repositioning strategies against SARS-CoV-2. Comput Struct Biotechnol J. 2022 Oct 17; 20: 5713-5728. doi: 10.1016/j.csbj.2022.10.017. Epub 2022 Oct 17. PMID: 36277237 [download]
- Maeda F, Kato A, Takeshima K, Shibazaki M, Sato R, Shibata T, Miyake K, Kozuka-Hata H, Oyama M, Shimizu E, Imoto S, Miyano S, Adachi S, Natsume T, Takeuchi K, Maruzuru Y, Koyanagi N, Jun A, Kawaguchi Y. Role of the Orphan Transporter SLC35E1 in the Nuclear Egress of Herpes Simplex Virus 1. J Virol. 2022 May 25; 96(10):e0030622. doi: 10.1128/jvi.00306-22. Epub 2022 Apr 27. PMID: 35475666 [download]
- Matsui Y, Abe Y, Uno K, Miyano S. RoDiCE: robust differential protein co-expression analysis for cancer complexome. Bioinformatics. 2022 Mar 1; 38(5): 1269–1276. DOI: 10.1093/bioinformatics/btab612. PMID: 34529752 [download]
- Mikami T, Kato I, Wing JB, Ueno H, Tasaka K, Tanaka K, Kubota H, Saida S, Umeda K, Hiramatsu H, Isobe T, Hiwatari M, Okada A, Chiba K, Shiraishi Y, Tanaka H, Miyano S, Arakawa Y, Oshima K, Koh K, Adachi S, Iwaisako K, Ogawa S, Sakaguchi S, Takita J. Alteration of the immune environment in bone marrow from children with recurrent B cell precursor acute lymphoblastic leukemia. Cancer Sci. 2022 Jan;113(1):41-52. doi: 10.1111/cas.15186. Epub 2021 Nov 29. PMID: 34716967 [download]
- Morita Y, Nannya Y, Ichikawa M, Hanamoto H, Shibayama H, Maeda Y, Hata T, Miyamoto T, Kawabata H, Takeuchi K, Tanaka H, Kishimoto J, Miyano S, Matsumura I, Ogawa S, Akashi K, Kanakura Y, Mitani K. ASXL1 mutations with serum EPO levels predict poor response to darbepoetin alfa in lower-risk MDS: W-JHS MDS01 trial. Int J Hematol. 2022 Nov;116(5):659-668. doi: 10.1007/s12185-022-03414-9. Epub 2022 Jul 12. PMID: 35821550 [download]
- Nakagawara K, Chubachi S, Namkoong H, Tanaka H, Lee H, Azekawa S, Otake S, Fukushima T, Morita A, Watase M, Sakurai K, Kusumoto T, Asakura T, Masaki K, Kamata H, Ishii M, Hasegawa N, Harada N, Ueda T, Ueda S, Ishiguro T, Arimura K, Saito F, Yoshiyama T, Nakano Y, Mutoh Y, Suzuki Y, Edahiro R, Murakami K, Sato Y, Okada Y, Koike R, Kitagawa Y, Tokunaga K, Kimura A, Imoto S, Miyano S, Ogawa S, Kanai T, Fukunaga K. Impact of upper and lower respiratory symptoms on COVID-19 outcomes: a multicenter retrospective cohort study. Respir Res. 2022 Nov 15;23(1):315. doi: 10.1186/s12931-022-02222-3. PMID: 36380316. [download]
- Namkoong H, Edahiro R, Takano T, Nishihara H, Shirai Y, Sonehara K, Tanaka H, Azekawa S, Mikami Y, Lee H, Hasegawa T, Okudela K, Okuzaki D, Motooka D, Kanai M, Naito T, Yamamoto K, Wang QS, Saiki R, Ishihara R, Matsubara Y, Hamamoto J, Hayashi H, Yoshimura Y, Tachikawa N, Yanagita E, Hyugaji T, Shimizu E, Katayama K, Kato Y, Morita T, Takahashi K, Harada N, Naito T, Hiki M, Matsushita Y, Takagi H, Aoki R, Nakamura A, Harada S, Sasano H, Kabata H, Masaki K, Kamata H, Ikemura S, Chubachi S, Okamori S, Terai H, Morita A, Asakura T, Sasaki J, Morisaki H, Uwamino Y, Nanki K, Uchida S, Uno S, Nishimura T, Ishiguro T, Isono T, Shibata S, Matsui Y, Hosoda C, Takano K, Nishida T, Kobayashi Y, Takaku Y, Takayanagi N, Ueda S, Tada A, Miyawaki M, Yamamoto M, Yoshida E, Hayashi R, Nagasaka T, Arai S, Kaneko Y, Sasaki K, Tagaya E, Kawana M, Arimura K, Takahashi K, Anzai T, Ito S, Endo A, Uchimura Y, Miyazaki Y, Honda T, Tateishi T, Tohda S, Ichimura N, Sonobe K, Sassa CT, Nakajima J, Nakano Y, Nakajima Y, Anan R, Arai R, Kurihara Y, Harada Y, Nishio K, Ueda T, Azuma M, Saito R, Sado T, Miyazaki Y, Sato R, Haruta Y, Nagasaki T, Yasui Y, Hasegawa Y, Mutoh Y, Kimura T, Sato T, Takei R, Hagimoto S, Noguchi Y, Yamano Y, Sasano H, Ota S, Nakamori Y, Yoshiya K, Saito F, Yoshihara T, Wada D, Iwamura H, Kanayama S, Maruyama S, Yoshiyama T, Ohta K, Kokuto H, Ogata H, Tanaka Y, Arakawa K, Shimoda M, Osawa T, Tateno H, Hase I, Yoshida S, Suzuki S, Kawada M, Horinouchi H, Saito F, Mitamura K, Hagihara M, Ochi J, Uchida T, Baba R, Arai D, Ogura T, Takahashi H, Hagiwara S, Nagao G, Konishi S, Nakachi I, Murakami K, Yamada M, Sugiura H, Sano H, Matsumoto S, Kimura N, Ono Y, Baba H, Suzuki Y, Nakayama S, Masuzawa K, Namba S, Suzuki K, Naito Y, Liu YC, Takuwa A, Sugihara F, Wing JB, Sakakibara S, Hizawa N, Shiroyama T, Miyawaki S, Kawamura Y, Nakayama A, Matsuo H, Maeda Y, Nii T, Noda Y, Niitsu T, Adachi Y, Enomoto T, Amiya S, Hara R, Yamaguchi Y, Murakami T, Kuge T, Matsumoto K, Yamamoto Y, Yamamoto M, Yoneda M, Kishikawa T, Yamada S, Kawabata S, Kijima N, Takagaki M, Sasa N, Ueno Y, Suzuki M, Takemoto N, Eguchi H, Fukusumi T, Imai T, Fukushima M, Kishima H, Inohara H, Tomono K, Kato K, Takahashi M, Matsuda F, Hirata H, Takeda Y, Koh H, Manabe T, Funatsu Y, Ito F, Fukui T, Shinozuka K, Kohashi S, Miyazaki M, Shoko T, Kojima M, Adachi T, Ishikawa M, Takahashi K, Inoue T, Hirano T, Kobayashi K, Takaoka H, Watanabe K, Miyazawa N, Kimura Y, Sado R, Sugimoto H, Kamiya A, Kuwahara N, Fujiwara A, Matsunaga T, Sato Y, Okada T, Hirai Y, Kawashima H, Narita A, Niwa K, Sekikawa Y, Nishi K, Nishitsuji M, Tani M, Suzuki J, Nakatsumi H, Ogura T, Kitamura H, Hagiwara E, Murohashi K, Okabayashi H, Mochimaru T, Nukaga S, Satomi R, Oyamada Y, Mori N, Baba T, Fukui Y, Odate M, Mashimo S, Makino Y, Yagi K, Hashiguchi M, Kagyo J, Shiomi T, Fuke S, Saito H, Tsuchida T, Fujitani S, Takita M, Morikawa D, Yoshida T, Izumo T, Inomata M, Kuse N, Awano N, Tone M, Ito A, Nakamura Y, Hoshino K, Maruyama J, Ishikura H, Takata T, Odani T, Amishima M, Hattori T, Shichinohe Y, Kagaya T, Kita T, Ohta K, Sakagami S, Koshida K, Hayashi K, Shimizu T, Kozu Y, Hiranuma H, Gon Y, Izumi N, Nagata K, Ueda K, Taki R, Hanada S, Kawamura K, Ichikado K, Nishiyama K, Muranaka H, Nakamura K, Hashimoto N, Wakahara K, Sakamoto K, Omote N, Ando A, Kodama N, Kaneyama Y, Maeda S, Kuraki T, Matsumoto T, Yokote K, Nakada TA, Abe R, Oshima T, Shimada T, Harada M, Takahashi T, Ono H, Sakurai T, Shibusawa T, Kimizuka Y, Kawana A, Sano T, Watanabe C, Suematsu R, Sageshima H, Yoshifuji A, Ito K, Takahashi S, Ishioka K, Nakamura M, Masuda M, Wakabayashi A, Watanabe H, Ueda S, Nishikawa M, Chihara Y, Takeuchi M, Onoi K, Shinozuka J, Sueyoshi A, Nagasaki Y, Okamoto M, Ishihara S, Shimo M, Tokunaga Y, Kusaka Y, Ohba T, Isogai S, Ogawa A, Inoue T, Fukuyama S, Eriguchi Y, Yonekawa A, Kan-O K, Matsumoto K, Kanaoka K, Ihara S, Komuta K, Inoue Y, Chiba S, Yamagata K, Hiramatsu Y, Kai H, Asano K, Oguma T, Ito Y, Hashimoto S, Yamasaki M, Kasamatsu Y, Komase Y, Hida N, Tsuburai T, Oyama B, Takada M, Kanda H, Kitagawa Y, Fukuta T, Miyake T, Yoshida S, Ogura S, Abe S, Kono Y, Togashi Y, Takoi H, Kikuchi R, Ogawa S, Ogata T, Ishihara S, Kanehiro A, Ozaki S, Fuchimoto Y, Wada S, Fujimoto N, Nishiyama K, Terashima M, Beppu S, Yoshida K, Narumoto O, Nagai H, Ooshima N, Motegi M, Umeda A, Miyagawa K, Shimada H, Endo M, Ohira Y, Watanabe M, Inoue S, Igarashi A, Sato M, Sagara H, Tanaka A, Ohta S, Kimura T, Shibata Y, Tanino Y, Nikaido T, Minemura H, Sato Y, Yamada Y, Hashino T, Shinoki M, Iwagoe H, Takahashi H, Fujii K, Kishi H, Kanai M, Imamura T, Yamashita T, Yatomi M, Maeno T, Hayashi S, Takahashi M, Kuramochi M, Kamimaki I, 1. Tominaga Y, Ishii T, Utsugi M, Ono A, Tanaka T, Kashiwada T, Fujita K, Saito Y, Seike M, Watanabe H, Matsuse H, Kodaka N, Nakano C, Oshio T, Hirouchi T, Makino S, Egi M; Biobank Japan Project, Omae Y, Nannya Y, Ueno T, Katayama K, Ai M, Fukui Y, Kumanogoh A, Sato T, Hasegawa N, Tokunaga K, Ishii M, Koike R, Kitagawa Y, Kimura A, Imoto S, Miyano S, Ogawa S, Kanai T, Fukunaga K, Okada Y. DOCK2 is involved in the host genetics and biology of severe COVID-19. Nature. 2022 Sep 22; 609(7928):754-760. doi: 10.1038/s41586-022-05163-5. Epub 2022 Aug 8. PMID: 35940203 [download]
- Ogasawara T, Fujii Y, Kakiuchi N, Shiozawa Y, Sakamoto R, Ogawa Y, Ootani K, Ito E, Tanaka T, Watanabe K, Yoshida Y, Kimura N, Shiraishi Y, Chiba K, Tanaka H, Miyano S, Ogawa S. Genetic Analysis of Pheochromocytoma and Paraganglioma Complicating Cyanotic Congenital Heart Disease. J Clin Endocrinol Metab. 2022 Aug 18; 107(9):2545-2555. doi: 10.1210/clinem/dgac362. PMID: 35730597 [download]
- Otake S, Chubachi S, Namkoong H, Nakagawara K, Tanaka H, Lee H, Morita A, Fukushima T, Watase M, Kusumoto T, Masaki K, Kamata H, Ishii M, Hasegawa N, Harada N, Ueda T, Ueda S, Ishiguro T, Arimura K, Saito F, Yoshiyama T, Nakano Y, Mutoh Y, Suzuki Y, Murakami K, Okada Y, Koike R, Kitagawa Y, Kimura A, Imoto S, Miyano S, Ogawa S, Kanai T, Fukunaga K; Japan COVID-19 Task Force. Clinical clustering with prognostic implications in Japanese COVID-19 patients: report from Japan COVID-19 Task Force, a nation-wide consortium to investigate COVID-19 host genetics. BMC Infect Dis. 2022 Sep 14;22(1):735. doi: 10.1186/s12879-022-07701-y. PMID: 36104674 [download]
- Park H, Imoto S, Miyano S. PredictiveNetwork: predictive gene network estimation with application to gastric cancer drug response-predictive network analysis. BMC Bioinformatics. 2022 Aug 16; 23(1):342. doi: 10.1186/s12859-022-04871-z. PMID: 35974335 [download]
- Park H, Miyano S. Computational Tactics for Precision Cancer Network Biology. Int J Mol Sci. 2022 Nov 19; 23(22):14398. doi: 10.3390/ijms232214398. PMID: 36430875 Review. https://www.mdpi.com/1422-0067/23/22/14398 [download]
- Park H, Yamaguchi R, Imoto S, Miyano S. Uncovering Molecular Mechanisms of Drug Resistance via Network-Constrained Common Structure Identification. J Comput Biol. 2022 Mar 9; 29(3):257-275. doi: 10.1089/cmb.2021.0314. Epub 2022 Jan 21. PMID: 35073162 [download]
- Park H, Yamaguchi R, Imoto S, Miyano S. Xprediction: Explainable EGFR-TKIs response prediction based on drug sensitivity specific gene networks. PLoS One. 2022 May 18; 17(5): e0261630. doi: 10.1371/journal.pone.0261630. eCollection 2022. PMID: 35584089 [download]
- Takeda J, Yoshida K, Nakagawa MM, Nannya Y, Yoda A, Saiki R, Ochi Y, Zhao L, Okuda R, Qi X, Mori T, Kon A, Chiba K, Tanaka H, Shiraishi Y, Kuo MC, Kerr CM, Nagata Y, Morishita D, Hiramoto N, Hangaishi A, Nakazawa H, Ishiyama K, Miyano S, Chiba S, Miyazaki Y, Kitano T, Usuki K, Sezaki N, Tsurumi H, Miyawaki S, Maciejewski JP, Ishikawa T, Ohyashiki K, Ganser A, Heuser M, Thol F, Shih LY, Takaori-Kondo A, Makishima H, Ogawa S. Amplified EPOR/JAK2 Genes Define a Unique Subtype of Acute Erythroid Leukemia. Blood Cancer Discov. 2022 Sep 6; (5):410-427. doi: 10.1158/2643-3230.BCD-21-0192. PMID: 35839275 [download]
- Takeda R, Yokoyama K, Fukuyama T, Kawamata T, Ito M, Yusa N, Kasajima R, Shimizu E, Ohno N, Uchimaru K, Yamaguchi R, Imoto S, Miyano S, Tojo A. Repeated Lineage Switches in an Elderly Case of Refractory B-Cell Acute Lymphoblastic Leukemia With MLL Gene Amplification: A Case Report and Literature Review. Front Oncol. 2022 Mar 23; 12: 799982. doi: 10.3389/fonc.2022.799982. PMID: 35402256 [download]
- Takeuchi Y, Yoshida K, Halik A, Kunitz A, Suzuki H, Kakiuchi N, Shiozawa Y, Yokoyama A, Inoue Y, Hirano T, Yoshizato T, Aoki K, Fujii Y, Nannya Y, Makishima H, Pfitzner BM, Bullinger L, Hirata M, Jinnouchi K, Shiraishi Y, Chiba K, Tanaka H, Miyano S, Okamoto T, Haga H, Ogawa S, Damm F. The landscape of genetic aberrations in myxofibrosarcoma. Int J Cancer. 2022 Aug 15; 151(4):565-577. doi: 10.1002/ijc.34051. Epub 2022 May 13. PMID: 35484982 [download]
- Wang QS, Edahiro R, Namkoong H, Hasegawa T, Shirai Y, Sonehara K, Tanaka H, Lee H, Saiki R, Hyugaji T, Shimizu E, Katayama K, Kanai M, Naito T, Sasa N, Yamamoto K, Kato Y, Morita T, Takahashi K, Harada N, Naito T, Hiki M, Matsushita Y, Takagi H, Ichikawa M, Nakamura A, Harada S, Sandhu Y, Kabata H, Masaki K, Kamata H, Ikemura S, Chubachi S, Okamori S, Terai H, Morita A, Asakura T, Sasaki J, Morisaki H, Uwamino Y, Nanki K, Uchida S, Uno S, Nishimura T, Ishiguro T, Isono T, Shibata S, Matsui Y, Hosoda C, Takano K, Nishida T, Kobayashi Y, Takaku Y, Takayanagi N, Ueda S, Tada A, Miyawaki M, Yamamoto M, Yoshida E, Hayashi R, Nagasaka T, Arai S, Kaneko Y, Sasaki K, Tagaya E, Kawana M, Arimura K, Takahashi K, Anzai T, Ito S, Endo A, Uchimura Y, Miyazaki Y, Honda T, Tateishi T, Tohda S, Ichimura N, Sonobe K, Sassa CT, Nakajima J, Nakano Y, Nakajima Y, Anan R, Arai R, Kurihara Y, Harada Y, Nishio K, Ueda T, Azuma M, Saito R, Sado T, Miyazaki Y, Sato R, Haruta Y, Nagasaki T, Yasui Y, Hasegawa Y, Mutoh Y, Kimura T, Sato T, Takei R, Hagimoto S, Noguchi Y, Yamano Y, Sasano H, Ota S, Nakamori Y, Yoshiya K, Saito F, Yoshihara T, Wada D, Iwamura H, Kanayama S, Maruyama S, Yoshiyama T, Ohta K, Kokuto H, Ogata H, Tanaka Y, Arakawa K, Shimoda M, Osawa T, Tateno H, Hase I, Yoshida S, Suzuki S, Kawada M, Horinouchi H, Saito F, Mitamura K, Hagihara M, Ochi J, Uchida T, Baba R, Arai D, Ogura T, Takahashi H, Hagiwara S, Nagao G, Konishi S, Nakachi I, Murakami K, Yamada M, Sugiura H, Sano H, Matsumoto S, Kimura N, Ono Y, Baba H, Suzuki Y, Nakayama S, Masuzawa K, Namba S, Shiroyama T, Noda Y, Niitsu T, Adachi Y, Enomoto T, Amiya S, Hara R, Yamaguchi Y, Murakami T, Kuge T, Matsumoto K, Yamamoto Y, Yamamoto M, Yoneda M, Tomono K, Kato K, Hirata H, Takeda Y, Koh H, Manabe T, Funatsu Y, Ito F, Fukui T, Shinozuka K, Kohashi S, Miyazaki M, Shoko T, Kojima M, Adachi T, Ishikawa M, Takahashi K, Inoue T, Hirano T, Kobayashi K, Takaoka H, Watanabe K, Miyazawa N, Kimura Y, Sado R, Sugimoto H, Kamiya A, Kuwahara N, Fujiwara A, Matsunaga T, Sato Y, Okada T, Hirai Y, Kawashima H, Narita A, Niwa K, Sekikawa Y, Nishi K, Nishitsuji M, Tani M, Suzuki J, Nakatsumi H, Ogura T, Kitamura H, Hagiwara E, Murohashi K, Okabayashi H, Mochimaru T, Nukaga S, Satomi R, Oyamada Y, Mori N, Baba T, Fukui Y, Odate M, Mashimo S, Makino Y, Yagi K, Hashiguchi M, Kagyo J, Shiomi T, Fuke S, Saito H, Tsuchida T, Fujitani S, Takita M, Morikawa D, Yoshida T, Izumo T, Inomata M, Kuse N, Awano N, Tone M, Ito A, Nakamura Y, Hoshino K, Maruyama J, Ishikura H, Takata T, Odani T, Amishima M, Hattori T, Shichinohe Y, Kagaya T, Kita T, Ohta K, Sakagami S, Koshida K, Hayashi K, Shimizu T, Kozu Y, Hiranuma H, Gon Y, Izumi N, Nagata K, Ueda K, Taki R, Hanada S, Kawamura K, Ichikado K, Nishiyama K, Muranaka H, Nakamura K, Hashimoto N, Wakahara K, Koji S, Omote N, Ando A, Kodama N, Kaneyama Y, Maeda S, Kuraki T, Matsumoto T, Yokote K, Nakada TA, Abe R, Oshima T, Shimada T, Harada M, Takahashi T, Ono H, Sakurai T, Shibusawa T, Kimizuka Y, Kawana A, Sano T, Watanabe C, Suematsu R, Sageshima H, Yoshifuji A, Ito K, Takahashi S, Ishioka K, Nakamura M, Masuda M, Wakabayashi A, Watanabe H, Ueda S, Nishikawa M, Chihara Y, Takeuchi M, Onoi K, Shinozuka J, Sueyoshi A, Nagasaki Y, Okamoto M, Ishihara S, Shimo M, Tokunaga Y, Kusaka Y, Ohba T, Isogai S, Ogawa A, Inoue T, Fukuyama S, Eriguchi Y, Yonekawa A, Kan-O K, Matsumoto K, Kanaoka K, Ihara S, Komuta K, Inoue Y, Chiba S, Yamagata K, Hiramatsu Y, Kai H, Asano K, Oguma T, Ito Y, Hashimoto S, Yamasaki M, Kasamatsu Y, Komase Y, Hida N, Tsuburai T, Oyama B, Takada M, Kanda H, Kitagawa Y, Fukuta T, Miyake T, Yoshida S, Ogura S, Abe S, Kono Y, Togashi Y, Takoi H, Kikuchi R, Ogawa S, Ogata T, Ishihara S, Kanehiro A, Ozaki S, Fuchimoto Y, Wada S, Fujimoto N, Nishiyama K, Terashima M, Beppu S, Yoshida K, Narumoto O, Nagai H, Ooshima N, Motegi M, Umeda A, Miyagawa K, Shimada H, Endo M, Ohira Y, Watanabe M, Inoue S, Igarashi A, Sato M, Sagara H, Tanaka A, Ohta S, Kimura T, Shibata Y, Tanino Y, Nikaido T, Minemura H, Sato Y, Yamada Y, Hashino T, Shinoki M, Iwagoe H, Takahashi H, Fujii K, Kishi H, Kanai M, Imamura T, Yamashita T, Yatomi M, Maeno T, Hayashi S, Takahashi M, Kuramochi M, Kamimaki I, Tominaga Y, Ishii T, Utsugi M, Ono A, Tanaka T, Kashiwada T, Fujita K, Saito Y, Seike M, Watanabe H, Matsuse H, Kodaka N, Nakano C, Oshio T, Hirouchi T, Makino S, Egi M, Omae Y, Nannya Y, Ueno T, Takano T, Katayama K, Ai M, Kumanogoh A, Sato T, Hasegawa N, Tokunaga K, Ishii M, Koike R, Kitagawa Y, Kimura A, Imoto S, Miyano S, Ogawa S, Kanai T, Fukunaga K, Okada Y. The whole blood transcriptional regulation landscape in 465 COVID-19 infected samples from Japan COVID-19 Task Force. Nat Commun. 2022 Aug 22; 13(1):4830. doi: 10.1038/s41467-022-32276-2. PMID: 35995775 [download]
- Watanabe K, Kimura S, Seki M, Isobe T, Kubota Y, Sekiguchi M, Sato-Otsubo A, Hiwatari M, Kato M, Oka A, Koh K, Sato Y, Tanaka H, Miyano S, Kawai T, Hata K, Ueno H, Nannya Y, Suzuki H, Yoshida K, Fujii Y, Nagae G, Aburatani H, Ogawa S, Takita J. Identification of the ultrahigh-risk subgroup in neuroblastoma cases through DNA methylation analysis and its treatment exploiting cancer metabolism. Oncogene. 2022 Nov 01; 41(46):4994-5007. doi: 10.1038/s41388-022-02489-2. Epub 2022 Nov 1. PMID: 36319669. https://www.nature.com/articles/s41388-022-02489-2 [download]
- Yaguchi T, Kimura S, Sekiguchi M, Kubota Y, Seki M, Yoshida K, Shiraishi Y, Kataoka K, Fujii Y, Watanabe K, Hiwatari M, Miyano S, Ogawa S, Takita J. Description of longitudinal tumor evolution in a case of multiply relapsed clear cell sarcoma of the kidney. Cancer Rep (Hoboken). 2022 Feb; 5(2):e1458. doi: 10.1002/cnr2.1458. Online ahead of print. PMID: 34967151 [download]
- Yamato G, Kawai T, Shiba N, Ikeda J, Hara Y, Ohki K, Tsujimoto SI, Kaburagi T, Yoshida K, Shiraishi Y, Miyano S, Kiyokawa N, Tomizawa D, Shimada A, Sotomatsu M, Arakawa H, Adachi S, Taga T, Horibe K, Ogawa S, Hata K, Hayashi Y. Genome-wide DNA methylation analysis in pediatric acute myeloid leukemia. Blood Adv. 2022 Jun 14; 6(11):3207-3219. doi: 10.1182/bloodadvances.2021005381. PMID: 35008106 [download]
- Zhang YZ, Yamaguchi K, Hatakeyama S, Furukawa Y, Miyano S, Yamaguchi R, Imoto S. On the application of BERT models for nanopore methylation detection. 2021 IEEE International Conference on Bioinformatics and Biomedicine (BIBM2021). 2021 Dec 9-12; 1: 320-327. https://www.computer.org/csdl/proceedings-article/bibm/2021/09669841/1A9Wo26lWj6 [download]
- Niida A, Mimori K, Shibata T, Miyano S. Modeling colorectal cancer evolution. J Hum Genet. 2021 Sep; 66 (9): 869-878. Review. doi: 10.1038/s10038-021-00930-0. Epub ahead of print 2021 May 13. PMID: 33986478. [download].
- Zhang YZ, Imoto S, Miyano S, Yamaguchi R. Enhancing breakpoint resolution with deep segmentation model: A general refinement method for read-depth based structural variant callers. PLoS Comput Biol. 2021 Oct 11;17(10):e1009186. doi: 10.1371/journal.pcbi.1009186. PMID: 34634042 [download]
- Yamaguchi K, Kasajima R, Takane K, Hatakeyama S, Shimizu E, Yamaguchi R, Katayama K, Arai M, Ishioka C, Iwama T, Kaneko S, Matsubara N, Moriya Y, Nomizu T, Sugano K, Tamura K, Tomita N, Yoshida T, Sugihara K, Nakamura Y, Miyano S, Imoto S, Furukawa Y, Ikenoue T. Application of targeted nanopore sequencing for the screening and determination of structural variants in patients with Lynch syndrome. J Hum Genet. 2021 Nov; 66(11):1053-1060. doi: 10.1038/s10038-021-00927-9. Epub 2021 May 6. PMID: 33958709 [download]
- Yada E, Kasajima R, Niida A, Imoto S, Miyano S, Miyagi Y, Sasada T, Wada S. Possible Role of Cytochrome P450 1B1 in the Mechanism of Gemcitabine Resistance in Pancreatic Cancer. Biomedicines. 2021 Oct 5; 9(10):1396. doi: 10.3390/biomedicines9101396. PMID: 34680513 [download]
- Tanaka H, Lee H, Morita A, Namkoong H, Chubachi S, Kabata H, Kamata H, Ishii M, Hasegawa N, Harada N, Ueda T, Ueda S, Ishiguro T, Arimura K, Saito F, Yoshiyama T, Nakano Y, Mutoh Y, Suzuki Y, Murakami K, Okada Y, Koike R, Kitagawa Y, Tokunaga K, Kimura A, Imoto S, Miyano S, Ogawa S, Kanai T, Fukunaga K; Japan COVID-19 Task Force. Clinical Characteristics of Patients with Coronavirus Disease (COVID-19): Preliminary Baseline Report of Japan COVID-19 Task Force, a Nationwide Consortium to Investigate Host Genetics of COVID-19. Int J Infect Dis. 2021 Dec; 113: 74-81. doi: 10.1016/j.ijid.2021.09.070. Epub 2021 Sep 30. PMID: 34601141 [download]
- Suzuki M, Kasajima R, Yokose T, Ito H, Shimizu E, Hatakeyama S, Yokoyama K, Yamaguchi R, Furukawa Y, Miyano S, Imoto S, Yoshioka E, Washimi K, Okubo Y, Kawachi K, Sato S, Miyagi Y. Comprehensive molecular analysis of genomic profiles and PD-L1 expression in lung adenocarcinoma with a high-grade fetal adenocarcinoma component. Transl Lung Cancer Res. 2021 Mar;10(3):1292-1304. doi: 10.21037/tlcr-20-1158. [download]
- Shimada K, Yoshida K, Suzuki Y, Iriyama C, Inoue Y, Sanada M, Kataoka K, Yuge M, Takagi Y, Kusumoto S, Masaki Y, Ito T, Inagaki Y, Okamoto A, Kuwatsuka Y, Nakatochi M, Shimada S, Miyoshi H, Shiraishi Y, Chiba K, Tanaka H, Miyano S, Shiozawa Y, Nannya Y, Okabe A, Kohno K, Atsuta Y, Ohshima K, Nakamura S, Ogawa S, Tomita A, Kiyoi H. Frequent genetic alterations in immune checkpoint-related genes in intravascular large B-cell lymphoma. Blood. 2021 Mar 18; 137(11):1491-1502. doi: 10.1182/Blood.2020007245. [download]
- Saiki R, Momozawa Y, Nannya Y, Nakagawa MM, Ochi Y, Yoshizato T, Terao C, Kuroda Y, Shiraishi Y, Chiba K, Tanaka H, Niida A, Imoto S, Matsuda K, Morisaki M, Murakami Y, Kamatani Y, Matsuda S, Kubo M, Miyano S, Makishima H, Ogawa S. Combined landscape of single-nucleotide variants and copy number alterations in clonal hematopoiesis. Nat Med. 2021 July; 27 (7):1239–1249. DOI: 10.1038/s41591-021-01411-9. [download]
- Polprasert C, Takeuchi Y, Makishima H, Wudhikarn K, Kakiuchi N, Tangnuntachai N, Assanasen T, Sitthi W, Muhamad H, Lawasut P, Kongkiatkamon S, Bunworasate U, Izutsu K, Shiraishi Y, Chiba K, Tanaka H, Miyano S, Ogawa S, Yoshida K, Rojnuckarin P. Frequent mutations in HLA and related genes in extranodal NK/T cell lymphomas. Leuk Lymphoma. 2021 Jan; 62(1):95-103. doi: 10.1080/10428194.2020.1821011. [download]
- Ochi Y, Yoshida K, Huang YJ, Kuo MC, Nannya Y, Sasaki K, Mitani K, Hosoya N, Hiramoto N, Ishikawa T, Branford S, Shanmuganathan N, Ohyashiki K, Takahashi N, Takaku T, Tsuchiya S, Kanemura N, Nakamura N, Ueda Y, Yoshihara S, Bera R, Shiozawa Y, Zhao L, Takeda J, Watatani Y, Okuda R, Makishima H, Shiraishi Y, Chiba K, Tanaka H, Sanada M, Takaori-Kondo A, Miyano S, Ogawa S, Shih LY. Clonal evolution and clinical implications of genetic abnormalities in blastic transformation of chronic myeloid leukaemia. Nat Commun. 2021 May 14; 12(1):2833. doi: 10.1038/s41467-021-23097-w. [download]
- Nishimura A, Hirabayashi S, Hasegawa D, Yoshida K, Shiraishi Y, Ashiarai M, Hosoya Y, Fujiwara T, Harigae H, Miyano S, Ogawa S, Manabe A. Acquisition of monosomy 7 and a RUNX1 mutation in Pearson syndrome. Pediatr Blood Cancer. 2021 Feb; 68(2):e28799. doi: 10.1002/pbc.28799. [download]
- Nakamura F, Arai H, Nannya Y, Ichikawa M, Furuichi S, Nagasawa F, Takahashi W, Handa T, Nakamura Y, Tanaka H, Nakamura Y, Sasaki K, Miyano S, Ogawa S, Mitani K. Development of Philadelphia chromosome-negative acute myeloid leukemia with IDH2 and NPM1 mutations in a patient with chronic myeloid leukemia who showed a major molecular response to tyrosine kinase inhibitor therapy. Int J Hematol. 2021 Jun; 113(6):936-940. doi: 10.1007/s12185-020-03074-7. [download]
- Sato N, Kakuta M, Hasegawa T, Yamaguchi R, Uchino E, Murashita K, Nakaji S, Imoto S, Yanagita M, Okuno Y. Metagenomic profiling of the gut microbiome in early chronic kidney disease. Nephrol Dial Transplant. 2021 Aug 27;36:1675-1684. doi: 10.1093/ndt/gfaa122. PMID: 32869063.
- Mizuno S, Yamaguchi R, Hasegawa T, Hayashi S, Fujita M, Zhang F, Koh Y, Lee SY, Yoon SS, Shimizu E, Komura M, Fujimoto A, Nagai M, Kato M, Liang H, Miyano S, Zhang Z, Nakagawa H, Imoto S. Immunogenomic pan-cancer landscape reveals immune escape mechanisms and immunoediting histories. Sci Rep. 2021 Aug 3;11(1):15713. doi: 10.1038/s41598-021-95287-x. [download]
- Li C, Qin J, Kuroyanagi K, Lu L, Nagasaki M, Miyano S. High-speed parameter search of dynamic biological pathways from time-course transcriptomic profiles using high-level Petri net. Biosystems. 2021 Mar; 201: 104332. doi: 10.1016/j.biosystems.2020.104332. [download]
- Koyamaishi S, Kamio T, Kobayashi A, Sato T, Kudo K, Sasaki S, Kanezaki R, Hasegawa D, Muramatsu H, Takahashi Y, Sasahara Y, Hiramatsu H, Kakuda H, Tanaka M, Ishimura M, Nishi M, Ishiguro A, Yabe H, Sarashina T, Yamamoto M, Yuza Y, Hyakuna N, Yoshida K, Kanno H, Ohga S, Ohara A, Kojima S, Miyano S, Ogawa S, Toki T, Terui K, Ito E. Reduced-intensity conditioning is effective for hematopoietic stem cell transplantation in young pediatric patients with Diamond-Blackfan anemia. Bone Marrow Transplant. 2021 May; 56(5):1013-1020. doi: 10.1038/s41409-020-01056-1. Erratum in: Bone Marrow Transplant. 2021 May; 56(5):1218-1219. doi: 10.1038/s41409-020-01076-x. [download]
- Kimura S, Sekiguchi M, Watanabe K, Hiwatarai M, Seki M, Yoshida K, Isobe T, Shiozawa Y, Suzuki H, Hoshino N, Hayashi Y, Oka A, Miyano S, Ogawa S, Takita J. Association of high-risk neuroblastoma classification based on expression profiles with differentiation and metabolism. PLoS One. 2021 Jan 19; 16(1):e0245526. doi: 10.1371/journal.pone.0245526. [download]
- Ishida Y, Kakiuchi N, Yoshida K, Inoue Y, Irie H, Kataoka TR, Hirata M, Funakoshi T, Matsushita S, Hata H, Uchi H, Yamamoto Y, Fujisawa Y, Fujimura T, Saiki R, Takeuchi K, Shiraishi Y, Chiba K, Tanaka H, Otsuka A, Miyano S, Kabashima K, Ogawa S. Unbiased Detection of Driver Mutations in Extramammary Paget Disease. Clin Cancer Res. 2021 Mar 15; 27(6):1756-1765. doi: 10.1158/1078-0432.CCR-20-3205. [download]
- Hirata H, Niida A, Kakiuchi N, Uchi R, Sugimachi K, Masuda T, Saito T, Kageyama SI, Motomura Y, Ito S, Yoshitake T, Tsurumaru D, Nishimuta Y, Yokoyama A, Hasegawa T, Chiba K, Shiraishi Y, Du J, Miura F, Morita M, Toh Y, Hirakawa M, Shioyama Y, Ito T, Akimoto T, Miyano S, Shibata T, Mori M, Suzuki Y, Ogawa S, Ishigami K, Mimori K. The Evolving Genomic Landscape of Esophageal Squamous Cell Carcinoma Under Chemoradiotherapy. Cancer Res. 2021 Oct 1;81(19):4926-4938. doi: 10.1158/0008-5472.CAN-21-0653. Epub 2021 Aug 19. PMID: 34413060 [download]
- Fujimoto K, Kimura Y, Allegretti JR, Yamamoto M, Zhang YZ, Katayama K, Tremmel G, Kawaguchi Y, Shimohigoshi M, Hayashi T, Uematsu M, Yamaguchi K, Furukawa Y, Akiyama Y, Yamaguchi R, Crowe SE, Ernst PB, Miyano S, Kiyono H, Imoto S, Uematsu S. Functional Restoration of Bacteriomes and Viromes by Fecal Microbiota Transplantation. Gastroenterology. 2021 May 1;160(6):2089-2102.e12. doi: 10.1053/j.gastro.2021.02.013. [download]
- Fujii Y, Sato Y, Suzuki H, Kakiuchi N, Shinzato T, Lenis AT, Maekawa S, Yokoyama A, Takeuchi Y, Inoue Y, Ochi Y, Shiozawa Y, Aoki K, Yoshida K, Kataoka K, Nakagawa MM, Nannya Y, Makishima H, Miyakawa J, Kawai T, Morikawa T, Shiraishi Y, Chiba K, Tanaka H, Nagae G, Sanada M, Sugihara E, Sato TA, Nakagawa T, Fukayama M, Ushiku T, Aburatani H, Miyano S, Coleman JA, Homma Y, Solit DB, Kume H, Ogawa S. Molecular classification and diagnostics of upper urinary tract urothelial carcinoma. Cancer Cell. 2021 Jun 14; 39(6):793-809.e8. doi: 10.1016/j.ccell.2021.05.008. PMID: 34129823 [download]
- Ebata N, Fujita M, Sasagawa S, Maejima K, Okawa Y, Hatanaka Y, Mitsuhashi T, Oosawa-Tatsuguchi A, Tanaka H, Miyano S, Nakamura T, Hirano S, Nakagawa H. Molecular Classification and Tumor Microenvironment Characterization of Gallbladder Cancer by Comprehensive Genomic and Transcriptomic Analysis. Cancers (Basel). 2021 Feb 10;13(4):733. doi: 10.3390/cancers13040733. PMID: 33578820 [download]
- COVID-19 Host Genetics Initiative (author list: Japan Coronavirus Taskforce Admin Team Lead: Takanori Kanai, Satoru Miyano, Seishi Ogawa). Mapping the human genetic architecture of COVID-19. Nature. 2021 Dec; 600(7889):472-477. doi: 10.1038/s41586-021-03767-x. Epub 2021 Jul 8. PMID: 34237774 [download]
- Sakimura S, Nagayama S, Fukunaga M, Hu Q, Kitagawa A, Kobayashi Y, Hasegawa T, Noda M, Kouyama Y, Shimizu D, Saito T, Niida A, Tsuruda Y, Otsu H, Matsumoto Y, Uchida H, Masuda T, Sugimachi K, Sasaki S, Yamada K, Takahashi K, Innan H, Suzuki Y, Nakamura H, Totoki Y, Mizuno S, Ohshima M, Shibata T, Mimori K. Impaired tumor immune response in metastatic tumors is a selective pressure for neutral evolution in colorectal cancer cases. PLoS Genet. 2021 Jan 21;17:e1009113. doi: 10.1371/journal.pgen.1009113. PMID: 33476333; PMCID: PMC7864431.
- Hasegawa T, Yamaguchi R, Kakuta M, Ando M, Songee J, Tokuda I, Murashita K, Imoto S. Application of a state-space model with skew-t measurement noise to blood-test value prediction. Appl Math Model. 2021;100:365-378. doi: 10.1016/j.apm.2021.08.007.
- Dingler FA, Wang M, Mu A, Millington CL, Oberbeck N, Watcham S, Pontel LB, Kamimae-Lanning AN, Langevin F, Nadler C, Cordell RL, Monks PS, Yu R, Wilson NK, Hira A, Yoshida K, Mori M, Okamoto Y, Okuno Y, Muramatsu H, Shiraishi Y, Kobayashi M, Moriguchi T, Osumi T, Kato M, Miyano S, Ito E, Kojima S, Yabe H, Yabe M, Matsuo K, Ogawa S, Göttgens B, Hodskinson MRG, Takata M, Patel KJ. Two Aldehyde Clearance Systems Are Essential to Prevent Lethal Formaldehyde Accumulation in Mice and Humans. Mol Cell. 2020 Dec 17; 80(6):996-1012.e9. doi: 10.1016/j.molcel.2020.10.012. [download]
- Imoto S, Hasegawa T, Yamaguchi R. Data science and precision health care. Nutr Rev. 2020 Dec 1;78(12 Suppl 2):53-57. doi: 10.1093/nutrit/nuaa110. PMID: 33259624.
- Moriyama T, Imoto S, Miyano S, Yamaguchi R. Theoretical Foundation of the Performance of Phylogeny-Based Somatic Variant Detection. Lecture Notes in Bioinformatics. 2020 Dec 7; 12508:87-101. Doi: 10.1007/978-3-030-64511-3_9 [download]
- Kudo SE, Kouyama Y, Ogawa Y, Ichimasa K, Hamada T, Kato K, Kudo K, Masuda T, Otsu H, Misawa M, Mori Y, Kudo T, Hayashi T, Wakamura K, Miyachi H, Sawada N, Sato T, Shibata T, Hamatani S, Nemoto T, Ishida F, Niida A, Miyano S, Oshima M, Ogino S, Mimori K. Depressed Colorectal Cancer: A New Paradigm in Early Colorectal Cancer. Clin Transl Gastroenterol. 2020 Dec; 11(12):e00269. doi: 10.14309/ctg.0000000000000269. [download]
- Kanamori T, Sanada M, Ri M, Ueno H, Nishijima D, Yasuda T, Tachita T, Narita T, Kusumoto S, Inagaki A, Ishihara R, Murakami Y, Kobayashi N, Shiozawa Y, Yoshida K, Nakagawa MM, Nannya Y, Shiraishi Y, Chiba K, Tanaka H, Miyano S, Horibe K, Handa H, Ogawa S, Iida S. Genomic analysis of multiple myeloma using targeted capture sequencing in the Japanese cohort. Br J Haematol. 2020 Dec; 191(5):755-763. doi: 10.1111/bjh.16720. [download]
- Ogura H, Ohga S, Aoki T, Utsugisawa T, Takahashi H, Iwai A, Watanabe K, Okuno Y, Yoshida K, Ogawa S, Miyano S, Kojima S, Yamamoto T, Yamamoto-Shimojima K, Kanno H. Novel COL4A1 mutations identified in infants with congenital hemolytic anemia in association with brain malformations. Hum Genome Var. 2020 Nov 27; 7(1):42. doi: 10.1038/s41439-020-00130-w. [download]
- Ishida S, Kato K, Tanaka M, Odamaki T, Kubo R, Mitsuyama E, Xiao JZ, Yamaguchi R, Uematsu S, Imoto S, Miyano S. Genome-wide association studies and heritability analysis reveal the involvement of host genetics in the Japanese gut microbiota. Commun Biol. 2020 Nov 18; 3(1):686. doi: 10.1038/s42003-020-01416-z. [download]
- Park H, Maruhashi K, Yamaguchi R, Imoto S, Miyano S. Global gene network exploration based on explainable artificial intelligence approach. PLoS One. 2020 Nov 6; 15(11):e0241508. doi: 10.1371/journal.pone.0241508. Erratum in: PLoS One. 2021 Jan 27; 16(1):e0246380. doi: 10.1371/journal.pone.0246380. [download]
- Hijikata Y, Yokoyama K, Yokoyama N, Matsubara Y, Shimizu E, Nakashima M, Yamagishi M, Ota Y, Lim LA, Yamaguchi R, Ito M, Tanaka Y, Denda T, Tani K, Yotsuyanagi H, Imoto S, Miyano S, Uchimaru K, Tojo A. Successful Clinical Sequencing by Molecular Tumor Board in an Elderly Patient With Refractory Sézary Syndrome. JCO Precis Oncol. 2020 Nov;4:534-560. doi: 10.1200/PO.19.00254. PMID: 35050744 [download]
- Ueno H, Yoshida K, Shiozawa Y, Nannya Y, Iijima-Yamashita Y, Kiyokawa N, Shiraishi Y, Chiba K, Tanaka H, Isobe T, Seki M, Kimura S, Makishima H, Nakagawa MM, Kakiuchi N, Kataoka K, Yoshizato T, Nishijima D, Deguchi T, Ohki K, Sato A, Takahashi H, Hashii Y, Tokimasa S, Hara J, Kosaka Y, Kato K, Inukai T, Takita J, Imamura T, Miyano S, Manabe A, Horibe K, Ogawa S, Sanada M. Landscape of driver mutations and their clinical impacts in pediatric B-cell precursor acute lymphoblastic leukemia. Blood Adv. 2020 Oct 27; 4(20):5165-5173. doi: 10.1182/Bloodadvances.2019001307. [download]
- Inagaki-Kawata Y, Yoshida K, Kawaguchi-Sakita N, Kawashima M, Nishimura T, Senda N, Shiozawa Y, Takeuchi Y, Inoue Y, Sato-Otsubo A, Fujii Y, Nannya Y, Suzuki E, Takada M, Tanaka H, Shiraishi Y, Chiba K, Kataoka Y, Torii M, Yoshibayashi H, Yamagami K, Okamura R, Moriguchi Y, Kato H, Tsuyuki S, Yamauchi A, Suwa H, Inamoto T, Miyano S, Ogawa S, Toi M. Genetic and clinical landscape of breast cancers with germline BRCA1/2 variants. Commun Biol. 2020 Oct 16; 3(1):578. doi: 10.1038/s42003-020-01301-9. [download]
- Matsuo H, Yoshida K, Nakatani K, Harata Y, Higashitani M, Ito Y, Kamikubo Y, Shiozawa Y, Shiraishi Y, Chiba K, Tanaka H, Okada A, Nannya Y, Takeda J, Ueno H, Kiyokawa N, Tomizawa D, Taga T, Tawa A, Miyano S, Meggendorfer M, Haferlach C, Ogawa S, Adachi S. Fusion partner-specific mutation profiles and KRAS mutations as adverse prognostic factors in MLL-rearranged AML. Blood Adv. 2020 Oct 13; 4(19):4623-4631. doi: 10.1182/Bloodadvances.2020002457. [download]
- Kubota Y, Seki M, Kawai T, Isobe T, Yoshida M, Sekiguchi M, Kimura S, Watanabe K, Sato-Otsubo A, Yoshida K, Suzuki H, Kataoka K, Fujii Y, Shiraishi Y, Chiba K, Tanaka H, Hiwatari M, Oka A, Hayashi Y, Miyano S, Ogawa S, Hata K, Tanaka Y, Takita J. Comprehensive genetic analysis of pediatric germ cell tumors identifies potential drug targets. Commun Biol. 2020 Sep 30; 3(1):544. doi: 10.1038/s42003-020-01267-8. [download]
- Hasegawa T, Hayashi S, Shimizu E, Mizuno S, Niida A, Yamaguchi R, Miyano S, Nakagawa H, Imoto S. Neoantimon: a multifunctional R package for identification of tumor-specific neoantigens. Bioinformatics. 2020 Sep 15; 36(18):4813-4816. doi: 10.1093/Bioinformatics/btaa616. [download]
- Fujimoto K, Kimura Y, Shimohigoshi M, Satoh T, Sato S, Tremmel G, Uematsu M, Kawaguchi Y, Usui Y, Nakano Y, Hayashi T, Kashima K, Yuki Y, Yamaguchi K, Furukawa Y, Kakuta M, Akiyama Y, Yamaguchi R, Crowe SE, Ernst PB, Miyano S, Kiyono H, Imoto S, Uematsu S. Metagenome Data on Intestinal Phage-Bacteria Associations Aids the Development of Phage Therapy against Pathobionts. Cell Host Microbe. 2020 Sep 9; 28(3):380-389.e9. doi: 10.1016/j.chom.2020.06.005. [download]
- Sekiguchi M, Seki M, Kawai T, Yoshida K, Yoshida M, Isobe T, Hoshino N, Shirai R, Tanaka M, Souzaki R, Watanabe K, Arakawa Y, Nannya Y, Suzuki H, Fujii Y, Kataoka K, Shiraishi Y, Chiba K, Tanaka H, Shimamura T, Sato Y, Sato-Otsubo A, Kimura S, Kubota Y, Hiwatari M, Koh K, Hayashi Y, Kanamori Y, Kasahara M, Kohashi K, Kato M, Yoshioka T, Matsumoto K, Oka A, Taguchi T, Sanada M, Tanaka Y, Miyano S, Hata K, Ogawa S, Takita J. Integrated multiomics analysis of hepatoblastoma unravels its heterogeneity and provides novel druggable targets. NPJ Precis Oncol. 2020 Jul 7; 4:20. doi: 10.1038/s41698-020-0125-y. [download]
- Maruhashi K, Park H, Yamaguchi R, Miyano S. Linear Tensor Projection Revealing Nonlinearity. CoRR. 2020 Jul; arXiv:2007.03912v1. [download]
- Dutta M, Nakagawa H, Kato H, Maejima K, Sasagawa S, Nakano K, Sasaki-Oku A, Fujimoto A, Mateos RN, Patil A, Tanaka H, Miyano S, Yasuda T, Nakai K, Fujita M. Whole genome sequencing analysis identifies recurrent structural alterations in esophageal squamous cell carcinoma. PeerJ. 2020 Jun 26; 8:e9294. doi: 10.7717/PeerJ.9294. [download]
- Ochi Y, Kon A, Sakata T, Nakagawa MM, Nakazawa N, Kakuta M, Kataoka K, Koseki H, Nakayama M, Morishita D, Tsuruyama T, Saiki R, Yoda A, Okuda R, Yoshizato T, Yoshida K, Shiozawa Y, Nannya Y, Kotani S, Kogure Y, Kakiuchi N, Nishimura T, Makishima H, Malcovati L, Yokoyama A, Takeuchi K, Sugihara E, Sato TA, Sanada M, Takaori-Kondo A, Cazzola M, Kengaku M, Miyano S, Shirahige K, Suzuki HI, Ogawa S. Combined Cohesin-RUNX1 Deficiency Synergistically Perturbs Chromatin Looping and Causes Myelodysplastic Syndromes. Cancer Discov. 2020 Jun; 10(6):836-853. doi: 10.1158/2159-8290.CD-19-0982. [download]
- Zhang YZ, Akdemir A, Tremmel G, Imoto S, Miyano S, Shibuya T, Yamaguchi R. Nanopore basecalling from a perspective of instance segmentation. BMC Bioinformatics. 2020 Apr 23; 21(Suppl 3):136. doi: 10.1186/s12859-020-3459-0. [download]
- Misawa K, Hasegawa T, Mishima E, Jutabha P, Ouchi M, Kojima K, Kawai Y, Matsuo M, Anzai N, Nagasaki M. Contribution of rare variants of the SLC22A12 gene to the missing heritability of serum urate levels. Genetics. 2020 Apr;214:1079-1090. doi: 10.1534/genetics.119.303006. Epub 2020 Jan 31. PMID: 32005656; PMCID: PMC7153932.
- Sato N, Kakuta M, Hasegawa T, Yamaguchi R, Uchino E, Kobayashi W, Sawada K, Tamura Y, Tokuda I, Murashita K, Nakaji S, Imoto S, Yanagita M, Okuno Y. Metagenomic analysis of bacterial species in the tongue microbiome of current and never smokers. NPJ Biofilms Microbiomes. 2020 Mar 13;6:11. doi: 10.1038/s41522-020-0121-6. PMID: 32170059; PMCID: PMC7069950.
- Hasegawa T, Yamaguchi R, Kakuta M, Sawada K, Kawatani K, Murashita K, Nakaji S, Imoto S. Prediction of blood-test values under different lifestyle scenarios using time-series electronic health records. PLoS One. 2020 Mar 20;15:e0230172. doi: 10.1371/journal.pone.0230172. PMID: 32196517; PMCID: PMC7083324.
- Sato N, Kakuta M, Uchino E, Hasegawa T, Kojima R, Kobayashi W, Sawada K, Tamura Y, Tokuda I, Imoto S, Nakaji S, Murashita K, Yanagita M, Okuno Y. The relationship between cigarette smoking and the tongue microbiome in an East Asian population. J Oral Microbiol. 2020 Mar 25;12:1742527. doi: 10.1080/20002297.2020.1742527. PMID: 32341759; PMCID: PMC7170382.
- Saito Y, Koya J, Araki M, Kogure Y, Shingaki S, Tabata M, McClure MB, Yoshifuji K, Matsumoto S, Isaka Y, Tanaka H, Kanai T, Miyano S, Shiraishi Y, Okuno Y, Kataoka K. Landscape and function of multiple mutations within individual oncogenes. Nature. 2020 Jun; 582(7810):95-99. doi: 10.1038/s41586-020-2175-2. [download]
- Niida A, Hasegawa T, Innan H, Shibata T, Mimori K, Miyano S. A unified simulation model for understanding the diversity of cancer evolution. PeerJ. 2020 Apr 8; 8:e8842. doi: 10.7717/PeerJ.8842. [download]
- Kimura S, Seki M, Kawai T, Goto H, Yoshida K, Isobe T, Sekiguchi M, Watanabe K, Kubota Y, Nannya Y, Ueno H, Shiozawa Y, Suzuki H, Shiraishi Y, Ohki K, Kato M, Koh K, Kobayashi R, Deguchi T, Hashii Y, Imamura T, Sato A, Kiyokawa N, Manabe A, Sanada M, Mansour MR, Ohara A, Horibe K, Kobayashi M, Oka A, Hayashi Y, Miyano S, Hata K, Ogawa S, Takita J. DNA methylation-based classification reveals difference between pediatric T-cell acute lymphoblastic leukemia and normal thymocytes. Leukemia. 2020 Apr; 34(4):1163-1168. doi: 10.1038/s41375-019-0626-2. [download]
- Fujimoto A, Fujita M, Hasegawa T, Wong JH, Maejima K, Oku-Sasaki A, Nakano K, Shiraishi Y, Miyano S, Yamamoto G, Akagi K, Imoto S, Nakagawa H. Comprehensive analysis of indels in whole-genome microsatellite regions and microsatellite instability across 21 cancer types. Genome Res. 2020 Mar 24; 30(3):334–346. doi: 10.1101/gr.255026.119. [download]
- Shrestha R, Sakata-Yanagimoto M, Maie K, Oshima M, Ishihara M, Suehara Y, Fukumoto K, Nakajima-Takagi Y, Matsui H, Kato T, Muto H, Sakamoto T, Kusakabe M, Nannya Y, Makishima H, Ueno H, Saiki R, Ogawa S, Chiba K, Shiraishi Y, Miyano S, Mouly E, Bernard OA, Inaba T, Koseki H, Iwama A, Chiba S. Molecular pathogenesis of progression to myeloid leukemia from TET-insufficient status. Blood Adv. 2020 Mar 10; 4(5):845-854. doi: 10.1182/Bloodadvances.2019001324. [download]
- Fujita M, Yamaguchi R, Hasegawa T, Shimada S, Arihiro K, Hayashi S, Maejima K, Nakano K, Fujimoto A, Ono A, Aikata H, Ueno M, Hayami S, Tanaka H, Miyano S, Yamaue H, Chayama K, Kakimi K, Tanaka S, Imoto S, Nakagawa H. Classification of primary liver cancer with immunosuppression mechanisms and correlation with genomic alterations. EBioMedicine. 2020 Mar; 53:102659. doi: 10.1016/j.ebiom.2020.102659. [download]
- Kasajima R, Yamaguchi R, Shimizu E, Tamada Y, Niida A, Tremmel G, Kishida T, Aoki I, Imoto S, Miyano S, Uemura H, Miyagi Y. Variant analysis of prostate cancer in Japanese patients and a new attempt to predict related biological pathways. Oncol Rep. 2020 Mar; 43(3):943-952. doi: 10.3892/or.2020.7481. [download]
- Hasegawa T, Yamaguchi R, Niida A, Miyano S, Imoto S. Ensemble smoothers for inference of hidden states and parameters in combinatorial regulatory model. J Frankl Inst. 2020 Mar; 357(5): 2916-2933. doi: 10.1016/j.jfranklin.2019.10.015. [download]
- Maeda-Minami A, Yoshino T, Katayama K, Horiba Y, Hikiami H, Shimada Y, Namiki T, Tahara E, Minamizawa K, Muramatsu S, Yamaguchi R, Imoto S, Miyano S, Mima H, Mimura M, Nakamura T, Watanabe K. Discrimination of prediction models between cold-heat and deficiency-excess patterns. Complement Ther Med. 2020 Mar; 49:102353. doi: 10.1016/j.ctim.2020.102353. [download]
- Taguchi M, Mishima H, Shiozawa Y, Hayashida C, Kinoshita A, Nannya Y, Makishima H, Horai M, Matsuo M, Sato S, Itonaga H, Kato T, Taniguchi H, Imanishi D, Imaizumi Y, Hata T, Takenaka M, Moriuchi Y, Shiraishi Y, Miyano S, Ogawa S, Yoshiura KI, Miyazaki Y. Genome analysis of myelodysplastic syndromes among atomic bomb survivors in Nagasaki. Haematologica. 2020 Jan 31; 105(2):358-365. doi: 10.3324/haematol.2019.219386. [download]
- Kakiuchi N, Yoshida K, Uchino M, Kihara T, Akaki K, Inoue Y, Kawada K, Nagayama S, Yokoyama A, Yamamoto S, Matsuura M, Horimatsu T, Hirano T, Goto N, Takeuchi Y, Ochi Y, Shiozawa Y, Kogure Y, Watatani Y, Fujii Y, Kim SK, Kon A, Kataoka K, Yoshizato T, Nakagawa MM, Yoda A, Nanya Y, Makishima H, Shiraishi Y, Chiba K, Tanaka H, Sanada M, Sugihara E, Sato TA, Maruyama T, Miyoshi H, Taketo MM, Oishi J, Inagaki R, Ueda Y, Okamoto S, Okajima H, Sakai Y, Sakurai T, Haga H, Hirota S, Ikeuchi H, Nakase H, Marusawa H, Chiba T, Takeuchi O, Miyano S, Seno H, Ogawa S. Frequent mutations that converge on the NFKBIZ pathway in ulcerative colitis. Nature. 2020 Jan; 577(7789):260-265. doi: 10.1038/s41586-019-1856-1. [download]
- PCAWG Consortium: Kadir C Akdemir, Eva G Alvarez, Adrian Baez-Ortega, Rameen Beroukhim, Paul C Boutros, David D L Bowtell, Benedikt Brors, Kathleen H Burns, Peter J Campbell, Kin Chan, Ken Chen, Isidro Cortés-Ciriano, Ana Dueso-Barroso, Andrew J Dunford, Paul A Edwards, Xavier Estivill, Dariush Etemadmoghadam, Lars Feuerbach, J Lynn Fink, Milana Frenkel-Morgenstern, Dale W Garsed, Mark Gerstein, Dmitry A Gordenin, David Haan, James E Haber, Julian M Hess, Barbara Hutter, Marcin Imielinski, David T W Jones, Young Seok Ju, Marat D Kazanov, Leszek J Klimczak, Youngil Koh, Jan O Korbel, Kiran Kumar, Eunjung Alice Lee, Jake June-Koo Lee, Yilong Li, Andy G Lynch, Geoff Macintyre, Florian Markowetz, Iñigo Martincorena, Alexander Martinez-Fundichely, Matthew Meyerson, Satoru Miyano, Hidewaki Nakagawa, Fabio C P Navarro, Stephan Ossowski, Peter J Park, John V Pearson, Montserrat Puiggròs, Karsten Rippe, Nicola D Roberts, Steven A Roberts, Bernardo Rodriguez-Martin, Steven E Schumacher, Ralph Scully, Mark Shackleton, Nikos Sidiropoulos, Lina Sieverling, Chip Stewart, David Torrents, Jose M C Tubio, Izar Villasante, Nicola Waddell, Jeremiah A Wala, Joachim Weischenfeldt, Lixing Yang, Xiaotong Yao, Sung-Soo Yoon, Jorge Zamora, Cheng-Zhong Zhang
- Rheinbay E, Nielsen MM, Abascal F, Wala JA, Shapira O, Tiao G, Hornshøj H, Hess JM, Juul RI, Lin Z, Feuerbach L, Sabarinathan R, Madsen T, Kim J, Mularoni L, Shuai S, Lanzós A, Herrmann C, Maruvka YE, Shen C, Amin SB, Bandopadhayay P, Bertl J, Boroevich KA, Busanovich J, Carlevaro-Fita J, Chakravarty D, Chan CWY, Craft D, Dhingra P, Diamanti K, Fonseca NA, Gonzalez-Perez A, Guo Q, Hamilton MP, Haradhvala NJ, Hong C, Isaev K, Johnson TA, Juul M, Kahles A, Kahraman A, Kim Y, Komorowski J, Kumar K, Kumar S, Lee D, Lehmann KV, Li Y, Liu EM, Lochovsky L, Park K, Pich O, Roberts ND, Saksena G, Schumacher SE, Sidiropoulos N, Sieverling L, Sinnott-Armstrong N, Stewart C, Tamborero D, Tubio JMC, Umer HM, Uusküla-Reimand L, Wadelius C, Wadi L, Yao X, Zhang CZ, Zhang J, Haber JE, Hobolth A, Imielinski M, Kellis M, Lawrence MS, von Mering C, Nakagawa H, Raphael BJ, Rubin MA, Sander C, Stein LD, Stuart JM, Tsunoda T, Wheeler DA, Johnson R, Reimand J, Gerstein M, Khurana E, Campbell PJ, López-Bigas N; PCAWG Drivers and Functional Interpretation Working Group; PCAWG Structural Variation Working Group, Weischenfeldt J, Beroukhim R, Martincorena I, Pedersen JS, Getz G; PCAWG Consortium. Analyses of non-coding somatic drivers in 2,658 cancer whole genomes. Nature. 2020 Feb 5; 578(7793):102-111. doi: 10.1038/s41586-020-1965-x. [download]
- Li Y, Roberts ND, Wala JA, Shapira O, Schumacher SE, Kumar K, Khurana E, Waszak S, Korbel JO, Haber JE, Imielinski M; PCAWG Structural Variation Working Group, Weischenfeldt J, Beroukhim R, Campbell PJ; PCAWG Consortium. Patterns of somatic structural variation in human cancer genomes. Nature. 2020 Feb 5; 578(7793):112-121. doi: 10.1038/s41586-019-1913-9. [download]
- ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium. Pan-cancer analysis of whole genomes. Nature. 2020 Feb 5; 578(7793):82-93. doi: 10.1038/s41586-020-1969-6. [download]
- Cortés-Ciriano I, Lee JJ, Xi R, Jain D, Jung YL, Yang L, Gordenin D, Klimczak LJ, Zhang CZ, Pellman DS; PCAWG Structural Variation Working Group, Park PJ; PCAWG Consortium. Comprehensive analysis of chromothripsis in 2,658 human cancers using whole-genome sequencing. Nat Genet. 2020 Mar; 52(3):331-341. doi: 10.1038/s41588-019-0576-7. Epub 2020 Feb 5. Erratum in: Nat Genet. 2020 May 13, doi: 10.1038/s41588-020-0634-1. [download]
- Akdemir KC, Le VT, Chandran S, Li Y, Verhaak RG, Beroukhim R, Campbell PJ, Chin L, Dixon JR, Futreal PA; PCAWG Structural Variation Working Group; PCAWG Consortium. Disruption of chromatin folding domains by somatic genomic rearrangements in human cancer. Nat Genet. 2020 Mar; 52(3):294-305. doi: 10.1038/s41588-019-0564-y. [download]
- Rodriguez-Martin B, Alvarez EG, Baez-Ortega A, Zamora J, Supek F, Demeulemeester J, Santamarina M, Ju YS, Temes J, Garcia-Souto D, Detering H, Li Y, Rodriguez-Castro J, Dueso-Barroso A, Bruzos AL, Dentro SC, Blanco MG, Contino G, Ardeljan D, Tojo M, Roberts ND, Zumalave S, Edwards PAW, Weischenfeldt J, Puiggròs M, Chong Z, Chen K, Lee EA, Wala JA, Raine K, Butler A, Waszak SM, Navarro FCP, Schumacher SE, Monlong J, Maura F, Bolli N, Bourque G, Gerstein M, Park PJ, Wedge DC, Beroukhim R, Torrents D, Korbel JO, Martincorena I, Fitzgerald RC, Van Loo P, Kazazian HH, Burns KH; PCAWG Structural Variation Working Group, Campbell PJ, Tubio JMC; PCAWG Consortium. Pan-cancer analysis of whole genomes identifies driver rearrangements promoted by LINE-1 retrotransposition. Nat Genet. 2020 Mar; 52(3):306-319. doi: 10.1038/s41588-019-0562-0. [download]
- Yakneen S, Waszak SM; PCAWG Technical Working Group, Gertz M, Korbel JO; PCAWG Consortium. Butler enables rapid cloud-based analysis of thousands of human genomes. Nat Biotechnol. 2020 Mar; 38(3):288-292. doi: 10.1038/s41587-019-0360-3. Epub 2020 Feb 5. Erratum in: Nat Biotechnol. 2020 Feb 12; doi: 10.1038/s41587-020-0448-9. [download]
- Zhang Y, Chen F, Fonseca NA, He Y, Fujita M, Nakagawa H, Zhang Z, Brazma A; PCAWG Transcriptome Working Group; PCAWG Structural Variation Working Group, Creighton CJ; PCAWG Consortium. High-coverage whole-genome analysis of 1220 cancers reveals hundreds of genes deregulated by rearrangement-mediated cis-regulatory alterations. Nat Commun. 2020 Feb 5; 11(1):736. doi: 10.1038/s41467-019-13885-w. [download]
- Sieverling L, Hong C, Koser SD, Ginsbach P, Kleinheinz K, Hutter B, Braun DM, Cortés-Ciriano I, Xi R, Kabbe R, Park PJ, Eils R, Schlesner M; PCAWG-Structural Variation Working Group, Brors B, Rippe K, Jones DTW, Feuerbach L; PCAWG Consortium. Genomic footprints of activated telomere maintenance mechanisms in cancer. Nat Commun. 2020 Feb 5;11(1):733. doi: 10.1038/s41467-019-13824-9. [download]
- Tsuda Y, Hirata M, Katayama K, Motoi T, Matsubara D, Oda Y, Fujita M, Kobayashi H, Kawano H, Nishida Y, Sakai T, Okuma T, Goto T, Ogura K, Kawai A, Ae K, Anazawa U, Suehara Y, Iwata S, Miyano S, Imoto S, Shibata T, Nakagawa H, Yamaguchi R, Tanaka S, Matsuda K. Massively parallel sequencing of tenosynovial giant cell tumors reveals novel CSF1 fusion transcripts and novel somatic CBL mutations. Int J Cancer. 2019 Dec 15; 145(12):3276-3284. doi: 10.1002/ijc.32421. [download]
- Hirata M, Asano N, Katayama K, Yoshida A, Tsuda Y, Sekimizu M, Mitani S, Kobayashi E, Komiyama M, Fujimoto H, Goto T, Iwamoto Y, Naka N, Iwata S, Nishida Y, Hiruma T, Hiraga H, Kawano H, Motoi T, Oda Y, Matsubara D, Fujita M, Shibata T, Nakagawa H, Nakayama R, Kondo T, Imoto S, Miyano S, Kawai A, Yamaguchi R, Ichikawa H, Matsuda K. Integrated exome and RNA sequencing of dedifferentiated liposarcoma. Nat Commun. 2019 Dec 12; 10(1):5683. doi: 10.1038/s41467-019-13286-z. Erratum in: Nat Commun. 2020 Feb 19; 11(1):1024. doi: 10.1038/s41467-020-14680-8. [download]
- Ito S, Yadome M, Nishiki T, Ishiduki S, Inoue H, Yamaguchi R, Miyano S. Virtual Grid Engine: a simulated grid engine environment for large-scale supercomputers. BMC Bioinformatics. 2019 Dec 2; 20(Suppl 16):591. doi: 10.1186/s12859-019-3085-x. [download]
- Watatani Y, Sato Y, Miyoshi H, Sakamoto K, Nishida K, Gion Y, Nagata Y, Shiraishi Y, Chiba K, Tanaka H, Zhao L, Ochi Y, Takeuchi Y, Takeda J, Ueno H, Kogure Y, Shiozawa Y, Kakiuchi N, Yoshizato T, Nakagawa MM, Nanya Y, Yoshida K, Makishima H, Sanada M, Sakata-Yanagimoto M, Chiba S, Matsuoka R, Noguchi M, Hiramoto N, Ishikawa T, Kitagawa J, Nakamura N, Tsurumi H, Miyazaki T, Kito Y, Miyano S, Shimoda K, Takeuchi K, Ohshima K, Yoshino T, Ogawa S, Kataoka K. Molecular heterogeneity in peripheral T-cell lymphoma, not otherwise specified revealed by comprehensive genetic profiling. Leukemia. 2019 Dec; 33(12):2867-2883. doi: 10.1038/s41375-019-0473-1. [download]
- Nagata Y, Makishima H, Kerr CM, Przychodzen BP, Aly M, Goyal A, Awada H, Asad MF, Kuzmanovic T, Suzuki H, Yoshizato T, Yoshida K, Chiba K, Tanaka H, Shiraishi Y, Miyano S, Mukherjee S, LaFramboise T, Nazha A, Sekeres MA, Radivoyevitch T, Haferlach T, Ogawa S, Maciejewski JP. Invariant patterns of clonal succession determine specific clinical features of myelodysplastic syndromes. Nat Commun. 2019 Nov 26; 10(1):5386. doi: 10.1038/s41467-019-13001-y. [download]
- Moriyama T, Imoto S, Hayashi S, Shiraishi Y, Miyano S, Yamaguchi R. Accurate and Flexible Bayesian Mutation Call from Multi-regional Tumor Samples. Lecture Notes in Computer Science. 2019 Nov 19; 11826: 47-61. doi: 10.1007/978-3-030-35210-3_4. [download]
- Moriyama T, Imoto S, Hayashi S, Shiraishi Y, Miyano S, Yamaguchi R. A Bayesian model integration for mutation calling through data partitioning. Bioinformatics. 2019 Nov 1; 35(21):4247-4254. doi: 10.1093/Bioinformatics/btz233. [download]
- Takeda R, Yokoyama K, Ogawa M, Kawamata T, Fukuyama T, Kondoh K, Takei T, Nakamura S, Ito M, Yusa N, Shimizu E, Ohno N, Uchimaru K, Yamaguchi R, Imoto S, Miyano S, Tojo A. The first case of elderly TCF3-HLF-positive B-cell acute lymphoblastic leukemia. Leuk Lymphoma. 2019 Nov; 60(11):2821-2824. doi: 10.1080/10428194.2019.1602267. [download]
- Shiba N, Yoshida K, Hara Y, Yamato G, Shiraishi Y, Matsuo H, Okuno Y, Chiba K, Tanaka H, Kaburagi T, Takeuchi M, Ohki K, Sanada M, Okubo J, Tomizawa D, Taki T, Shimada A, Sotomatsu M, Horibe K, Taga T, Adachi S, Tawa A, Miyano S, Ogawa S, Hayashi Y. Transcriptome analysis offers a comprehensive illustration of the genetic background of pediatric acute myeloid leukemia. Blood Adv. 2019 Oct 22; 3(20):3157-3169. doi: 10.1182/Bloodadvances.2019000404. [download]
- Takeda R, Yokoyama K, Kobayashi S, Kawamata T, Nakamura S, Fukuyama T, Ito M, Yusa N, Shimizu E, Ohno N, Yamaguchi R, Imoto S, Miyano S, Uchimaru K, Tojo A. An Unusually Short Latent Period of Therapy-Related Myeloid Neoplasm Harboring a Rare MLL-EP300 Rearrangement: Case Report and Literature Review. Case Rep Hematol. 2019 Oct 2; 2019:4532434. doi: 10.1155/2019/4532434. [download]
- Osawa T, Shimamura T, Saito K, Hasegawa Y, Ishii N, Nishida M, Ando R, Kondo A, Anwar M, Tsuchida R, Hino S, Sakamoto A, Igarashi K, Saitoh K, Kato K, Endo K, Yamano S, Kanki Y, Matsumura Y, Minami T, Tanaka T, Anai M, Wada Y, Wanibuchi H, Hayashi M, Hamada A, Yoshida M, Yachida S, Nakao M, Sakai J, Aburatani H, Shibuya M, Hanada K, Miyano S, Soga T, Kodama T. Phosphoethanolamine Accumulation Protects Cancer Cells under Glutamine Starvation through Downregulation of PCYT2. Cell Rep. 2019 Oct 1; 29(1):89-103.e7. doi: 10.1016/j.celrep.2019.08.087. [download]
- Kubota Y, Uryu K, Ito T, Seki M, Kawai T, Isobe T, Kumagai T, Toki T, Yoshida K, Suzuki H, Kataoka K, Shiraishi Y, Chiba K, Tanaka H, Ohki K, Kiyokawa N, Kagawa J, Miyano S, Oka A, Hayashi Y, Ogawa S, Terui K, Sato A, Hata K, Ito E, Takita J. Integrated genetic and epigenetic analysis revealed heterogeneity of acute lymphoblastic leukemia in Down syndrome. Cancer Sci. 2019 Oct; 110(10):3358-3367. doi: 10.1111/cas.14160. [download]
- Mori M, Hira A, Yoshida K, Muramatsu H, Okuno Y, Shiraishi Y, Anmae M, Yasuda J, Tadaka S, Kinoshita K, Osumi T, Noguchi Y, Adachi S, Kobayashi R, Kawabata H, Imai K, Morio T, Tamura K, Takaori-Kondo A, Yamamoto M, Miyano S, Kojima S, Ito E, Ogawa S, Matsuo K, Yabe H, Yabe M, Takata M. Pathogenic mutations identified by a multimodality approach in 117 Japanese Fanconi anemia patients. Haematologica. 2019 Oct;104(10):1962-1973. doi: 10.3324/haematol.2018.207241. Erratum in: Haematologica. 2020 Apr; 105(4):1166-1167. doi: 10.3324/haematol.2019.245720. [download]
- Kajino T, Shimamura T, Gong S, Yanagisawa K, Ida L, Nakatochi M, Griesing S, Shimada Y, Kano K, Suzuki M, Miyano S, Takahashi T. Divergent lncRNA MYMLR regulates MYC by eliciting DNA looping and promoter-enhancer interaction. EMBO J. 2019 Sep 2; 38(17):e98441. doi: 10.15252/embj.201798441. [download]
- Matsuno Y, Atsumi Y, Shimizu A, Katayama K, Fujimori H, Hyodo M, Minakawa Y, Nakatsu Y, Kaneko S, Hamamoto R, Shimamura T, Miyano S, Tsuzuki T, Hanaoka F, Yoshioka KI. Replication stress triggers microsatellite destabilization and hypermutation leading to clonal expansion in vitro. Nat Commun. 2019 Sep 2; 10(1):3925. doi: 10.1038/s41467-019-11760-2. [download]
- Hayashi S, Moriyama T, Yamaguchi R, Mizuno S, Komura M, Miyano S, Nakagawa H, Imoto S. ALPHLARD-NT: Bayesian Method for Human Leukocyte Antigen Genotyping and Mutation Calling through Simultaneous Analysis of Normal and Tumor Whole-Genome Sequence Data. J Comput Biol. 2019 Sep; 26(9):923-937. doi: 10.1089/cmb.2018.0224. [download]
- Maeda-Minami A, Yoshino T, Katayama K, Horiba Y, Hikiami H, Shimada Y, Namiki T, Tahara E, Minamizawa K, Muramatsu S, Yamaguchi R, Imoto S, Miyano S, Mima H, Mimura M, Nakamura T, Watanabe K. Prediction of deficiency-excess pattern in Japanese Kampo medicine: Multi-centre data collection. Complement Ther Med. 2019 Aug; 45:228-233. doi: 10.1016/j.ctim.2019.07.003. [download]
- Mateos RN, Nakagawa H, Hirono S, Takano S, Fukasawa M, Yanagisawa A, Yasukawa S, Maejima K, Oku-Sasaki A, Nakano K, Dutta M, Tanaka H, Miyano S, Enomoto N, Yamaue H, Nakai K, Fujita M. Genomic analysis of pancreatic juice DNA assesses malignant risk of intraductal papillary mucinous neoplasm of pancreas. Cancer Med. 2019 Aug; 8(10):4565-4573. doi: 10.1002/cam4.2340. [download]
- Yamaguchi K, Shimizu E, Yamaguchi R, Imoto S, Komura M, Hatakeyama S, Noguchi R, Takane K, Ikenoue T, Gohda Y, Yano H, Miyano S, Furukawa Y. Development of an MSI-positive colon tumor with aberrant DNA methylation in a PPAP patient. J Hum Genet. 2019 Aug; 64(8):729-740. doi: 10.1038/s10038-019-0611-7. [download]
- Yoshino T, Katayama K, Yamaguchi R, Imoto S, Miyano S, Mima H, Watanabe K. Classification of patients with cold sensation by a review of systems database: A single-centre observational study. Complement Ther Med. 2019 Aug; 45:7-13. doi: 10.1016/j.ctim.2019.05.011. [download]
- Kataoka K, Miyoshi H, Sakata S, Dobashi A, Couronné L, Kogure Y, Sato Y, Nishida K, Gion Y, Shiraishi Y, Tanaka H, Chiba K, Watatani Y, Kakiuchi N, Shiozawa Y, Yoshizato T, Yoshida K, Makishima H, Sanada M, Onozawa M, Teshima T, Yoshiki Y, Ishida T, Suzuki K, Shimada K, Tomita A, Kato M, Ota Y, Izutsu K, Demachi-Okamura A, Akatsuka Y, Miyano S, Yoshino T, Gaulard P, Hermine O, Takeuchi K, Ohshima K, Ogawa S. Frequent structural variations involving programmed death ligands in Epstein-Barr virus-associated lymphomas. Leukemia. 2019 Jul; 33(7):1687-1699. doi: 10.1038/s41375-019-0380-5. [download]
- Nagao Y, Mimura N, Takeda J, Yoshida K, Shiozawa Y, Oshima M, Aoyama K, Saraya A, Koide S, Rizq O, Hasegawa Y, Shiraishi Y, Chiba K, Tanaka H, Nishijima D, Isshiki Y, Kayamori K, Kawajiri-Manako C, Oshima-Hasegawa N, Tsukamoto S, Mitsukawa S, Takeda Y, Ohwada C, Takeuchi M, Iseki T, Misawa S, Miyano S, Ohara O, Yokote K, Sakaida E, Kuwabara S, Sanada M, Iwama A, Ogawa S, Nakaseko C. Genetic and transcriptional landscape of plasma cells in POEMS syndrome. Leukemia. 2019 Jul; 33(7):1723-1735. doi: 10.1038/s41375-018-0348-x. [download]
- Kim SK, Takeda H, Takai A, Matsumoto T, Kakiuchi N, Yokoyama A, Yoshida K, Kaido T, Uemoto S, Minamiguchi S, Haga H, Shiraishi Y, Miyano S, Seno H, Ogawa S, Marusawa H. Comprehensive analysis of genetic aberrations linked to tumorigenesis in regenerative nodules of liver cirrhosis. J Gastroenterol. 2019 Jul; 54(7):628-640. doi: 10.1007/s00535-019-01555-z. [download]
- Nakamura S, Yokoyama K, Shimizu E, Yusa N, Kondoh K, Ogawa M, Takei T, Kobayashi A, Ito M, Isobe M, Konuma T, Kato S, Kasajima R, Wada Y, Nagamura-Inoue T, Yamaguchi R, Takahashi S, Imoto S, Miyano S, Tojo A. Prognostic impact of circulating tumor DNA status post-allogeneic hematopoietic stem cell transplantation in AML and MDS. Blood. 2019 Jun 20; 133(25):2682-2695. doi: 10.1182/Blood-2018-10-880690. [download]
- VanderWeele DJ, Finney R, Katayama K, Gillard M, Paner G, Imoto S, Yamaguchi R, Wheeler D, Lack J, Cam M, Pontier A, Nguyen YTM, Maejima K, Sasaki-Oku A, Nakano K, Tanaka H, Vander Griend D, Kubo M, Ratain MJ, Miyano S, Nakagawa H. Genomic Heterogeneity Within Individual Prostate Cancer Foci Impacts Predictive Biomarkers of Targeted Therapy. Eur Urol Focus. 2019 May;5(3):416-424. doi: 10.1016/j.euf.2018.01.006. [download]
- Niida A, Hasegawa T, Miyano S. Sensitivity analysis of agent-based simulation utilizing massively parallel computation and interactive data visualization. PLoS One. 2019 Mar 5; 14(3):e0210678. doi: 10.1371/journal.pone.0210678. [download]
- Muraoka D, Seo N, Hayashi T, Tahara Y, Fujii K, Tawara I, Miyahara Y, Okamori K, Yagita H, Imoto S, Yamaguchi R, Komura M, Miyano S, Goto M, Sawada SI, Asai A, Ikeda H, Akiyoshi K, Harada N, Shiku H. Antigen delivery targeted to tumor-associated macrophages overcomes tumor immune resistance. J Clin Invest. 2019 Mar 1; 129(3):1278-1294. doi: 10.1172/JCI97642. [download]
- Kimura S, Hasegawa D, Yoshimoto Y, Seki M, Daida A, Sekiguchi M, Hirabayashi S, Hosoya Y, Kobayashi M, Miyano S, Ogawa S, Takita J, Manabe A. Duplication of ALK F1245 missense mutation due to acquired uniparental disomy associated with aggressive progression in a patient with relapsed neuroblastoma. Oncol Lett. 2019 Mar; 17(3):3323-3329. doi: 10.3892/ol.2019.9985. [download]
- Okuno Y, Murata T, Sato Y, Muramatsu H, Ito Y, Watanabe T, Okuno T, Murakami N, Yoshida K, Sawada A, Inoue M, Kawa K, Seto M, Ohshima K, Shiraishi Y, Chiba K, Tanaka H, Miyano S, Narita Y, Yoshida M, Goshima F, Kawada JI, Nishida T, Kiyoi H, Kato S, Nakamura S, Morishima S, Yoshikawa T, Fujiwara S, Shimizu N, Isobe Y, Noguchi M, Kikuta A, Iwatsuki K, Takahashi Y, Kojima S, Ogawa S, Kimura H. Defective Epstein-Barr virus in chronic active infection and haematological malignancy. Nat Microbiol. 2019 Mar; 4(3):404-413. doi: 10.1038/s41564-018-0334-0. Erratum in: Nat Microbiol. 2019 Mar; 4(3):544. doi: 10.1038/s41564-019-0387-8. [download]
- Kotani S, Yoda A, Kon A, Kataoka K, Ochi Y, Shiozawa Y, Hirsch C, Takeda J, Ueno H, Yoshizato T, Yoshida K, Nakagawa MM, Nannya Y, Kakiuchi N, Yamauchi T, Aoki K, Shiraishi Y, Miyano S, Maeda T, Maciejewski JP, Takaori-Kondo A, Ogawa S, Makishima H. Molecular pathogenesis of disease progression in MLL-rearranged AML. Leukemia. 2019 Mar; 33(3):612-624. doi: 10.1038/s41375-018-0253-3. [download]
- Park H, Yamada M, Imoto S, Miyano S. Robust Sample-Specific Stability Selection with Effective Error Control. J Comput Biol. 2019 Mar; 26(3):202-217. doi: 10.1089/cmb.2018.0180. [download]
- Polprasert C, Takeuchi Y, Kakiuchi N, Yoshida K, Assanasen T, Sitthi W, Bunworasate U, Pirunsarn A, Wudhikarn K, Lawasut P, Uaprasert N, Kongkiatkamon S, Moonla C, Sanada M, Akita N, Takeda J, Fujii Y, Suzuki H, Nannya Y, Shiraishi Y, Chiba K, Tanaka H, Miyano S, Rojnuckarin P, Ogawa S, Makishima H. Frequent germline mutations of HAVCR2 in sporadic subcutaneous panniculitis-like T-cell lymphoma. Blood Adv. 2019 Feb 26; 3(4):588-595. doi: 10.1182/Bloodadvances.2018028340. [download]
- Matsui Y, Miyano S, Shimamura T. Tumor Subclonal Progression Model for Cancer Hallmark Acquisition. Lecture Notes in Bioinformatics. 2019 Feb 14; 10834: 115-123. doi: 10.1007/978-3-030-14160-8_12. [download]
- Shimamura T, Matsui Y, Kajino T, Ito S, Takahashi T, Miyano S. GIMLET: Identifying Biological Modulators in Context-Specific Gene Regulation Using Local Energy Statistics. Lecture Notes in Computer Science. 2019 Feb 14; 10834: 124-137. doi: 10.1007/978-3-030-14160-8_13. [download]
- Kimura S, Seki M, Yoshida K, Shiraishi Y, Akiyama M, Koh K, Imamura T, Manabe A, Hayashi Y, Kobayashi M, Oka A, Miyano S, Ogawa S, Takita J. NOTCH1 pathway activating mutations and clonal evolution in pediatric T-cell acute lymphoblastic leukemia. Cancer Sci. 2019 Feb; 110(2):784-794. doi: 10.1111/cas.13859. [download]
- Ono S, Matsuda J, Watanabe E, Akaike H, Teranishi H, Miyata I, Otomo T, Sadahira Y, Mizuochi T, Kusano H, Kage M, Ueno H, Yoshida K, Shiraishi Y, Chiba K, Tanaka H, Miyano S, Ogawa S, Hayashi Y, Kanegane H, Ouchi K. Novel neuroblastoma amplified sequence (NBAS) mutations in a Japanese boy with fever-triggered recurrent acute liver failure. Hum Genome Var. 2019 Jan 7; 6: 2. doi: 10.1038/s41439-018-0035-5. [download]
- Yokoyama A, Kakiuchi N, Yoshizato T, Nannya Y, Suzuki H, Takeuchi Y, Shiozawa Y, Sato Y, Aoki K, Kim SK, Fujii Y, Yoshida K, Kataoka K, Nakagawa MM, Inoue Y, Hirano T, Shiraishi Y, Chiba K, Tanaka H, Sanada M, Nishikawa Y, Amanuma Y, Ohashi S, Aoyama I, Horimatsu T, Miyamoto S, Tsunoda S, Sakai Y, Narahara M, Brown JB, Sato Y, Sawada G, Mimori K, Minamiguchi S, Haga H, Seno H, Miyano S, Makishima H, Muto M, Ogawa S. Age-related remodelling of oesophageal epithelia by mutated cancer drivers. Nature. 2019 Jan 2; 565(7739):312-317. doi: 10.1038/s41586-018-0811-x. [download]
- Hoshino A, Yang X, Tanita K, Yoshida K, Ono T, Nishida N, Okuno Y, Kanzaki T, Goi K, Fujino H, Ohshima K, Shiraishi Y, Chiba K, Tanaka H, Miyano S, Ogawa S, Kojima S, Morio T, Kanegane H. Modification of cellular and humoral immunity by somatically reverted T cells in X-linked lymphoproliferative syndrome type 1. J Allergy Clin Immunol. 2019 Jan 1; 143(1):421-424.e11. doi: 10.1016/j.jaci.2018.07.044. [download]
- Kobayashi K, Mizuta S, Yamane N, Ueno H, Yoshida K, Kato I, Umeda K, Hiramatsu H, Suehiro M, Maihara T, Usami I, Shiraishi Y, Chiba K, Miyano S, Adachi S, Ogawa S, Kiyokawa N, Heike T. Paraneoplastic hypereosinophilic syndrome associated with IL3-IgH positive acute lymphoblastic leukemia. Pediatr Blood Cancer. 2019 Jan; 66(1):e27449. doi: 10.1002/pbc.27449. [download]
- Aoki K, Nakamura H, Suzuki H, Matsuo K, Kataoka K, Shimamura T, Motomura K, Ohka F, Shiina S, Yamamoto T, Nagata Y, Yoshizato T, Mizoguchi M, Abe T, Momii Y, Muragaki Y, Watanabe R, Ito I, Sanada M, Yajima H, Morita N, Takeuchi I, Miyano S, Wakabayashi T, Ogawa S, Natsume A. Prognostic relevance of genetic alterations in diffuse lower-grade gliomas. Neuro Oncol. 2018 Jan 10;20(1):66-77. doi: 10.1093/neuonc/nox132. [download]
- Berger G, Kroeze LI, Koorenhof-Scheele TN, de Graaf AO, Yoshida K, Ueno H, Shiraishi Y, Miyano S, van den Berg E, Schepers H, van der Reijden BA, Ogawa S, Vellenga E, Jansen JH. Early detection and evolution of preleukemic clones in therapy-related myeloid neoplasms following autologous SCT. Blood. 2018 Apr 19;131(16):1846-1857. doi: 10.1182/Blood-2017-09-805879. [download]
- Hasegawa T, Kojima K, Kawai Y, Nagasaki M. Time-series filtering for replicated observations via a kernel approximate Bayesian computation. IEEE Trans Signal Process. 2018 Dec 1;66:6148-6161. doi: 10.1109/TSP.2018.2872864.
- Cardinez C, Miraghazadeh B, Tanita K, da Silva E, Hoshino A, Okada S, Chand R, Asano T, Tsumura M, Yoshida K, Ohnishi H, Kato Z, Yamazaki M, Okuno Y, Miyano S, Kojima S, Ogawa S, Andrews TD, Field MA, Burgio G, Morio T, Vinuesa CG, Kanegane H, Cook MC. Gain-of-function IKBKB mutation causes human combined immune deficiency. J Exp Med. 2018 Nov 5; 215(11):2715-2724. doi: 10.1084/jem.20180639. [download]
- Fujita K, Chen X, Homma H, Tagawa K, Amano M, Saito A, Imoto S, Akatsu H, Hashizume Y, Kaibuchi K, Miyano S, Okazawa H. Targeting Tyro3 ameliorates a model of PGRN-mutant FTLD-TDP via tau-mediated synaptic pathology. Nat Commun. 2018 Jan 30;9(1):433. doi: 10.1038/s41467-018-02821-z. [download]
- Furuta M, Tanaka H, Shiraishi Y, Unida T, Imamura M, Fujimoto A, Fujita M, Sasaki-Oku A, Maejima K, Nakano K, Kawakami Y, Arihiro K, Aikata H, Ueno M, Hayami S, Ariizumi SI, Yamamoto M, Gotoh K, Ohdan H, Yamaue H, Miyano S, Chayama K, Nakagawa H. Characterization of HBV integration patterns and timing in liver cancer and HBV-infected livers. Oncotarget. 2018 May 18; 9(38):25075-25088. doi: 10.18632/Oncotarget.25308. Erratum in: Oncotarget. 2018 Aug 3; 9(60):31789. doi: 10.18632/oncotarget.25960. [download]
- Hamada M, Doisaki S, Okuno Y, Muramatsu H, Hama A, Kawashima N, Narita A, Nishio N, Yoshida K, Kanno H, Manabe A, Taga T, Takahashi Y, Miyano S, Ogawa S, Kojima S. Whole-exome analysis to detect congenital hemolytic anemia mimicking congenital dyserythropoietic anemia. Int J Hematol. 2018 Sep; 108(3):306-311. doi: 10.1007/s12185-018-2482-7. [download]
- Hasegawa T, Kojima K, Kawai Y, Nagasaki M. Time-Series Filtering for Replicated Observations via a Kernel Approximate Bayesian Computation. IEEE Trans. Signal Process. 2018 Sep 30; 66(23): 6148-6161. DOI: 10.1109/TSP.2018.2872864
- Hayashi S, Yamaguchi R, Mizuno S, Komura M, Miyano S, Nakagawa H, Imoto S. ALPHLARD: a Bayesian method for analyzing HLA genes from whole genome sequence data. BMC Genomics. 2018 Nov 1; 19(1):790. doi: 10.1186/s12864-018-5169-9. [download]
- Hiramoto N, Takeda J, Yoshida K, Ono Y, Yoshioka S, Yamauchi N, Fujimoto A, Maruoka H, Shiraishi Y, Tanaka H, Chiba K, Imai Y, Miyano S, Ogawa S, Ishikawa T. Donor cell-derived transient abnormal myelopoiesis as a specific complication of umbilical cord Blood transplantation. Bone Marrow Transplant. 2018 Feb;53(2):225-227. doi: 10.1038/bmt.2017.226. [download]
- Hoshino A, Takashima T, Yoshida K, Morimoto A, Kawahara Y, Yeh TW, Okano T, Yamashita M, Mitsuiki N, Imai K, Sakatani T, Nakazawa A, Okuno Y, Shiraishi Y, Chiba K, Tanaka H, Miyano S, Ogawa S, Kojima S, Morio T, Kanegane H. Dysregulation of Epstein-Barr Virus Infection in Hypomorphic ZAP70 Mutation. J Infect Dis. 2018 Jul 24; 218(5):825-834. doi: 10.1093/infdis/jiy231. [download]
- Inoue D, Fujino T, Sheridan P, Zhang YZ, Nagase R, Horikawa S, Li Z, Matsui H, Kanai A, Saika M, Yamaguchi R, Kozuka-Hata H, Kawabata KC, Yokoyama A, Goyama S, Inaba T, Imoto S, Miyano S, Xu M, Yang FC, Oyama M, Kitamura T. A novel ASXL1-OGT axis plays roles in H3K4 methylation and tumor suppression in myeloid malignancies. Leukemia. 2018 Jun; 32(6):1327-1337. doi: 10.1038/s41375-018-0083-3. [download]
- Isobe T, Seki M, Yoshida K, Sekiguchi M, Shiozawa Y, Shiraishi Y, Kimura S, Yoshida M, Inoue Y, Yokoyama A, Kakiuchi N, Suzuki H, Kataoka K, Sato Y, Kawai T, Chiba K, Tanaka H, Shimamura T, Kato M, Iguchi A, Hama A, Taguchi T, Akiyama M, Fujimura J, Inoue A, Ito T, Deguchi T, Kiyotani C, Iehara T, Hosoi H, Oka A, Sanada M, Tanaka Y, Hata K, Miyano S, Ogawa S, Takita J. Integrated Molecular Characterization of the Lethal Pediatric Cancer Pancreatoblastoma. Cancer Res. 2018 Feb 15;78(4):865-876. doi: 10.1158/0008-5472.CAN-17-2581. [download]
- Ito S, Yadome M, Nishiki T, Ishiduki S, Inoue H, Yamaguchi R, Miyano S. Virtual Grid Engine: Accelerating thousands of omics sample analyses using large-scale supercomputers. 2018 IEEE International Conference on Bioinformatics and Biomedicine (BIBM). 2018 Dec; 387-392. doi: 10.1109/BIBM.2018.8621285. [download]
- Kamijo R, Itonaga H, Kihara R, Nagata Y, Hata T, Asou N, Ohtake S, Shiraishi Y, Chiba K, Tanaka H, Miyano S, Ogawa S, Naoe T, Kiyoi H, Miyazaki Y. Distinct gene alterations with a high percentage of myeloperoxidase-positive leukemic blasts in de novo acute myeloid leukemia. Leuk Res. 2018 Feb; 65:34-41. doi: 10.1016/j.leukres.2017.12.006. [download]
- Katagiri S, Umezu T, Azuma K, Asano M, Akahane D, Makishima H, Yoshida K, Watatani Y, Chiba K, Miyano S, Ogawa S, Ohyashiki JH, Ohyashiki K. Hidden FLT3-D835Y clone in FLT3-ITD-positive acute myeloid leukemia that evolved into very late relapse with T-lymphoblastic leukemia. Leuk Lymphoma. 2018 Jun; 59(6):1490-1493. doi: 10.1080/10428194.2017.1382696. [download]
- Kataoka K, Iwanaga M, Yasunaga JI, Nagata Y, Kitanaka A, Kameda T, Yoshimitsu M, Shiraishi Y, Sato-Otsubo A, Sanada M, Chiba K, Tanaka H, Ochi Y, Aoki K, Suzuki H, Shiozawa Y, Yoshizato T, Sato Y, Yoshida K, Nosaka K, Hishizawa M, Itonaga H, Imaizumi Y, Munakata W, Shide K, Kubuki Y, Hidaka T, Nakamaki T, Ishiyama K, Miyawaki S, Ishii R, Nureki O, Tobinai K, Miyazaki Y, Takaori-Kondo A, Shibata T, Miyano S, Ishitsuka K, Utsunomiya A, Shimoda K, Matsuoka M, Watanabe T, Ogawa S. Prognostic relevance of integrated genetic profiling in adult T-cell leukemia/lymphoma. Blood. 2018 Jan 11; 131(2):215-225. doi: 10.1182/Blood-2017-01-761874. [download]
- Kiyotani K, Mai TH, Yamaguchi R, Yew PY, Kulis M, Orgel K, Imoto S, Miyano S, Burks AW, Nakamura Y. Characterization of the B-cell receptor repertoires in peanut allergic subjects undergoing oral immunotherapy. J Hum Genet. 2018 Feb; 63(2):239-248. doi: 10.1038/s10038-017-0364-0. [download]
- Kobayashi M, Yokoyama K, Shimizu E, Yusa N, Ito M, Yamaguchi R, Imoto S, Miyano S, Tojo A. Phenotype-based gene analysis allowed successful diagnosis of X-linked neutropenia associated with a novel WASp mutation. Ann Hematol. 2018 Feb; 97(2):367-369. doi: 10.1007/s00277-017-3134-3. [download]
- Matsuo H, Yoshida K, Fukumura K, Nakatani K, Noguchi Y, Takasaki S, Noura M, Shiozawa Y, Shiraishi Y, Chiba K, Tanaka H, Okada A, Nannya Y, Takeda J, Ueno H, Shiba N, Yamato G, Handa H, Ono Y, Hiramoto N, Ishikawa T, Usuki K, Ishiyama K, Miyawaki S, Itonaga H, Miyazaki Y, Kawamura M, Yamaguchi H, Kiyokawa N, Tomizawa D, Taga T, Tawa A, Hayashi Y, Mano H, Miyano S, Kamikubo Y, Ogawa S, Adachi S. Recurrent CCND3 mutations in MLL-rearranged acute myeloid leukemia. Blood Adv. 2018 Nov 13; 2(21):2879-2889. doi: 10.1182/Bloodadvances.2018019398. [download]
- Mimori K, Saito T, Niida A, Miyano S. Cancer evolution and heterogeneity. Ann Gastroenterol Surg. 2018 Jul 4; 2(5):332-338. Review. doi: 10.1002/ags3.12182. [download]
- Murakami N, Okuno Y, Yoshida K, Shiraishi Y, Nagae G, Suzuki K, Narita A, Sakaguchi H, Kawashima N, Wang X, Xu Y, Chiba K, Tanaka H, Hama A, Sanada M, Ito M, Hirayama M, Watanabe A, Ueno T, Kojima S, Aburatani H, Mano H, Miyano S, Ogawa S, Takahashi Y, Muramatsu H. Integrated molecular profiling of juvenile myelomonocytic leukemia. Blood. 2018 Apr 5; 131(14):1576-1586. doi: 10.1182/Blood-2017-07-798157. [download]
- Nakamura S, Yokoyama K, Yusa N, Ogawa M, Takei T, Kobayashi A, Ito M, Shimizu E, Kasajima R, Wada Y, Yamaguchi R, Imoto S, Nagamura-Inoue T, Miyano S, Tojo A. Circulating tumor DNA dynamically predicts response and/or relapse in patients with hematological malignancies. Int J Hematol. 2018 Oct; 108(4):402-410. doi: 10.1007/s12185-018-2487-2. [download]
- Niida A, Nagayama S, Miyano S, Mimori K. Understanding intratumor heterogeneity by combining genome analysis and mathematical modeling. Cancer Sci. 2018 Apr; 109(4):884-892. doi: 10.1111/cas.13510. Erratum in: Cancer Sci. 2021 Jun;112(6):2556. doi: 10.1111/cas.14936. [download]
- Ochi Y, Hiramoto N, Yoshizato T, Ono Y, Takeda J, Shiozawa Y, Yoshida K, Kakiuchi N, Shiraishi Y, Tanaka H, Chiba K, Kazuma Y, Tabata S, Yonetani N, Uehara K, Yamashita D, Imai Y, Nagafuji K, Yamakawa M, Miyano S, Takaori-Kondo A, Ogawa S, Ishikawa T. Clonally related diffuse large B-cell lymphoma and interdigitating dendritic cell sarcoma sharing MYC translocation. Haematologica. 2018 Nov; 103(11):e553-e556. doi: 10.3324/haematol.2018.193490. [download]
- Ogawa M, Yokoyama K, Hirano M, Jimbo K, Ochi K, Kawamata T, Ohno N, Shimizu E, Yokoyama N, Yamaguchi R, Imoto S, Uchimaru K, Takahashi N, Miyano S, Imai Y, Tojo A. Different clonal dynamics of chronic myeloid leukaemia between bone marrow and the central nervous system. Br J Haematol. 2018 Dec; 183(5):842-845. doi: 10.1111/bjh.15065. [download]
- Park H, Shimamura T, Imoto S, Miyano S. Adaptive NetworkProfiler for Identifying Cancer Characteristic-Specific Gene Regulatory Networks. J Comput Biol. 2018 Feb; 25(2):130-145. doi: 10.1089/cmb.2017.0120. [download]
- Saito T, Niida A, Uchi R, Hirata H, Komatsu H, Sakimura S, Hayashi S, Nambara S, Kuroda Y, Ito S, Eguchi H, Masuda T, Sugimachi K, Tobo T, Nishida H, Daa T, Chiba K, Shiraishi Y, Yoshizato T, Kodama M, Okimoto T, Mizukami K, Ogawa R, Okamoto K, Shuto M, Fukuda K, Matsui Y, Shimamura T, Hasegawa T, Doki Y, Nagayama S, Yamada K, Kato M, Shibata T, Mori M, Aburatani H, Murakami K, Suzuki Y, Ogawa S, Miyano S, Mimori K. A temporal shift of the evolutionary principle shaping intratumor heterogeneity in colorectal cancer. Nat Commun. 2018 Jul 23; 9(1):2884. doi: 10.1038/s41467-018-05226-0. [download]
- Sakai H, Hosono N, Nakazawa H, Przychodzen B, Polprasert C, Carraway HE, Sekeres MA, Radivoyevitch T, Yoshida K, Sanada M, Yoshizato T, Kataoka K, Nakagawa MM, Ueno H, Nannya Y, Kon A, Shiozawa Y, Takeda J, Shiraishi Y, Chiba K, Miyano S, Singh J, Padgett RA, Ogawa S, Maciejewski JP, Makishima H. A novel genetic and morphologic phenotype of ARID2-mediated myelodysplasia. Leukemia. 2018 Mar; 32(3):839-843. doi: 10.1038/leu.2017.319. [download]
- Shiozawa Y, Malcovati L, Gallì A, Sato-Otsubo A, Kataoka K, Sato Y, Watatani Y, Suzuki H, Yoshizato T, Yoshida K, Sanada M, Makishima H, Shiraishi Y, Chiba K, Hellström-Lindberg E, Miyano S, Ogawa S, Cazzola M. Aberrant splicing and defective mRNA production induced by somatic spliceosome mutations in myelodysplasia. Nat Commun. 2018 Sep 7; 9(1):3649. doi: 10.1038/s41467-018-06063-x. [download]
- Shiraishi Y, Kataoka K, Chiba K, Okada A, Kogure Y, Tanaka H, Ogawa S, Miyano S. A comprehensive characterization of cis-acting splicing-associated variants in human cancer. Genome Res. 2018 Aug; 28(8):1111-1125. doi: 10.1101/gr.231951.117. [download]
- Takagi M, Hoshino A, Yoshida K, Ueno H, Imai K, Piao J, Kanegane H, Yamashita M, Okano T, Muramatsu H, Okuno Y, Shiraishi Y, Chiba K, Tanaka H, Miyano S, Ogawa S, Hayashi Y, Kojima S, Morio T. Genetic heterogeneity of uncharacterized childhood autoimmune diseases with lymphoproliferation. Pediatr Blood Cancer. 2018 Feb; 65(2): e26831. doi: 10.1002/pbc.26831. [download]
- Takei T, Yokoyama K, Shimizu E, Konuma T, Takahashi S, Yamaguchi R, Imoto S, Miyano S, Tojo A. Azacitidine effectively reduces TP53-mutant leukemic cell burden in secondary acute myeloid leukemia after cord blood transplantation. Leuk Lymphoma. 2018 Nov; 59(11):2755-2756. doi: 10.1080/10428194.2018.1443335. [download]
- Toki T, Yoshida K, Wang R, Nakamura S, Maekawa T, Goi K, Katoh MC, Mizuno S, Sugiyama F, Kanezaki R, Uechi T, Nakajima Y, Sato Y, Okuno Y, Sato-Otsubo A, Shiozawa Y, Kataoka K, Shiraishi Y, Sanada M, Chiba K, Tanaka H, Terui K, Sato T, Kamio T, Sakaguchi H, Ohga S, Kuramitsu M, Hamaguchi I, Ohara A, Kanno H, Miyano S, Kojima S, Ishiguro A, Sugita K, Kenmochi N, Takahashi S, Eto K, Ogawa S, Ito E. De Novo Mutations Activating Germline TP53 in an Inherited Bone-Marrow-Failure Syndrome. Am J Hum Genet. 2018 Sep 6; 103(3):440-447. doi: 10.1016/j.ajhg.2018.07.020. [download]
- Wardell CP, Fujita M, Yamada T, Simbolo M, Fassan M, Karlic R, Polak P, Kim J, Hatanaka Y, Maejima K, Lawlor RT, Nakanishi Y, Mitsuhashi T, Fujimoto A, Furuta M, Ruzzenente A, Conci S, Oosawa A, Sasaki-Oku A, Nakano K, Tanaka H, Yamamoto Y, Michiaki K, Kawakami Y, Aikata H, Ueno M, Hayami S, Gotoh K, Ariizumi SI, Yamamoto M, Yamaue H, Chayama K, Miyano S, Getz G, Scarpa A, Hirano S, Nakamura T, Nakagawa H. Genomic characterization of biliary tract cancers identifies driver genes and predisposing mutations. J Hepatol. 2018 May;68(5):959-969. doi: 10.1016/j.jhep.2018.01.009. [download]
- Yamato G, Shiba N, Yoshida K, Hara Y, Shiraishi Y, Ohki K, Okubo J, Park MJ, Sotomatsu M, Arakawa H, Kiyokawa N, Tomizawa D, Adachi S, Taga T, Horibe K, Miyano S, Ogawa S, Hayashi Y. RUNX1 mutations in pediatric acute myeloid leukemia are associated with distinct genetic features and an inferior prognosis. Blood. 2018 May 17; 131(20):2266-2270. doi: 10.1182/Blood-2017-11-814442. [download]
- Yokoyama K, Shimizu E, Yokoyama N, Nakamura S, Kasajima R, Ogawa M, Takei T, Ito M, Kobayashi A, Yamaguchi R, Imoto S, Miyano S, Tojo A. Cell-lineage level-targeted sequencing to identify acute myeloid leukemia with myelodysplasia-related changes. Blood Adv. 2018 Oct 9;2(19):2513-2521. doi: 10.1182/Bloodadvances.2017010744. [download]
- Anderson WP; Global Life Science Data Resources Working Group: Apweiler R, Bateman A, Bauer GA, Berman H, Blake JA, Blomberg N, Burley SK, Cochrane G, Di Francesco V, Donohue T, Durinx C, Game A, Green E, Gojobori T, Goodhand P, Hamosh A, Hermjakob H, Kanehisa M, Kiley R, McEntyre J, McKibbin R, Miyano S, Pauly B, Perrimon N, Ragan MA, Richards G, Teo YY, Westerfield M, Westhof E, Lasko PF. Data management: A global coalition to sustain core data. Nature. 2017 Mar 8;543(7644):179. doi: 10.1038/543179a. PMID: 28277502 [download]
- da Silva-Coelho P, Kroeze LI, Yoshida K, Koorenhof-Scheele TN, Knops R, van de Locht LT, de Graaf AO, Massop M, Sandmann S, Dugas M, Stevens-Kroef MJ, Cermak J, Shiraishi Y, Chiba K, Tanaka H, Miyano S, de Witte T, Blijlevens NMA, Muus P, Huls G, van der Reijden BA, Ogawa S, Jansen JH. Clonal evolution in myelodysplastic syndromes. Nat Commun. 2017 Apr 21; 8:15099. doi: 10.1038/ncomms15099. [download]
- Ding LW, Sun QY, Tan KT, Chien W, Mayakonda A, Yeoh AEJ, Kawamata N, Nagata Y, Xiao JF, Loh XY, Lin DC, Garg M, Jiang YY, Xu L, Lim SL, Liu LZ, Madan V, Sanada M, Fernández LT, Hema Preethi SS, Lill M, Kantarjian HM, Kornblau SM, Miyano S, Liang DC, Ogawa S, Shih LY, Yang H, Koeffler HP. Mutational Landscape of Pediatric Acute Lymphoblastic Leukemia. Cancer Res. 2017 Jan 15;77(2):390-400. doi: 10.1158/0008-5472.CAN-16-1303. Epub 2016 Nov 21. Erratum in: Cancer Res. 2017 Apr 15; 77(8):2174. doi: 10.1158/0008-5472.CAN-17-0404. [download]
- Fujii K, Miyahara Y, Harada N, Muraoka D, Komura M, Yamaguchi R, Yagita H, Nakamura J, Sugino S, Okumura S, Imoto S, Miyano S, Shiku H. Identification of an immunogenic neo-epitope encoded by mouse sarcoma using CXCR3 ligand mRNAs as sensors. Oncoimmunology. 2017 Mar 20; 6(5):e1306617. doi: 10.1080/2162402X.2017.1306617. [download]
- Fujita M, Matsubara N, Matsuda I, Maejima K, Oosawa A, Yamano T, Fujimoto A, Furuta M, Nakano K, Oku-Sasaki A, Tanaka H, Shiraishi Y, Mateos RN, Nakai K, Miyano S, Tomita N, Hirota S, Ikeuchi H, Nakagawa H. Genomic landscape of colitis-associated cancer indicates the impact of chronic inflammation and its stratification by mutations in the Wnt signaling. Oncotarget. 2017 Dec 12; 9(1):969-981. doi: 10.18632/Oncotarget.22867. [download]
- Furuta M, Ueno M, Fujimoto A, Hayami S, Yasukawa S, Kojima F, Arihiro K, Kawakami Y, Wardell CP, Shiraishi Y, Tanaka H, Nakano K, Maejima K, Sasaki-Oku A, Tokunaga N, Boroevich KA, Abe T, Aikata H, Ohdan H, Gotoh K, Kubo M, Tsunoda T, Miyano S, Chayama K, Yamaue H, Nakagawa H. Whole genome sequencing discriminates hepatocellular carcinoma with intrahepatic metastasis from multi-centric tumors. J Hepatol. 2017 Feb; 66(2):363-373. doi: 10.1016/j.jhep.2016.09.021. [download]
- Hirabayashi S, Seki M, Hasegawa D, Kato M, Hyakuna N, Shuo T, Kimura S, Yoshida K, Kataoka K, Fujii Y, Shiraishi Y, Chiba K, Tanaka H, Kiyokawa N, Miyano S, Ogawa S, Takita J, Manabe A. Constitutional abnormalities of IDH1 combined with secondary mutations predispose a patient with Maffucci syndrome to acute lymphoblastic leukemia. Pediatr Blood Cancer. 2017 Dec; 64(12): e26647. doi: 10.1002/pbc.26647. [download]
- Hiwatari M, Seki M, Akahoshi S, Yoshida K, Miyano S, Shiraishi Y, Tanaka H, Chiba K, Ogawa S, Takita J. Molecular studies reveal MLL-MLLT10/AF10 and ARID5B-MLL gene fusions displaced in a case of infantile acute lymphoblastic leukemia with complex karyotype. Oncol Lett. 2017 Aug;14(2):2295-2299. doi: 10.3892/ol.2017.6430. [download]
- Hoshino A, Okada S, Yoshida K, Nishida N, Okuno Y, Ueno H, Yamashita M, Okano T, Tsumura M, Nishimura S, Sakata S, Kobayashi M, Nakamura H, Kamizono J, Mitsui-Sekinaka K, Ichimura T, Ohga S, Nakazawa Y, Takagi M, Imai K, Shiraishi Y, Chiba K, Tanaka H, Miyano S, Ogawa S, Kojima S, Nonoyama S, Morio T, Kanegane H. Abnormal hematopoiesis and autoimmunity in human subjects with germline IKZF1 mutations. J Allergy Clin Immunol. 2017 Jul; 140(1):223-231. doi: 10.1016/j.jaci.2016.09.029. [download]
- Hosono N, Makishima H, Mahfouz R, Przychodzen B, Yoshida K, Jerez A, LaFramboise T, Polprasert C, Clemente MJ, Shiraishi Y, Chiba K, Tanaka H, Miyano S, Sanada M, Cui E, Verma AK, McDevitt MA, List AF, Saunthararajah Y, Sekeres MA, Boultwood J, Ogawa S, Maciejewski JP. Recurrent genetic defects on chromosome 5q in myeloid neoplasms. Oncotarget. 2017 Jan 24; 8(4):6483-6495. doi: 10.18632/Oncotarget.14130. [download]
- Ichimura T, Yoshida K, Okuno Y, Yujiri T, Nagai K, Nishi M, Shiraishi Y, Ueno H, Toki T, Chiba K, Tanaka H, Muramatsu H, Hara T, Kanno H, Kojima S, Miyano S, Ito E, Ogawa S, Ohga S. Diagnostic challenge of Diamond-Blackfan anemia in mothers and children by whole-exome sequencing. Int J Hematol. 2017 Apr; 105(4):515-520. doi: 10.1007/s12185-016-2151-7. [download]
- Ikeda F, Yoshida K, Toki T, Uechi T, Ishida S, Nakajima Y, Sasahara Y, Okuno Y, Kanezaki R, Terui K, Kamio T, Kobayashi A, Fujita T, Sato-Otsubo A, Shiraishi Y, Tanaka H, Chiba K, Muramatsu H, Kanno H, Ohga S, Ohara A, Kojima S, Kenmochi N, Miyano S, Ogawa S, Ito E. Exome sequencing identified RPS15A as a novel causative gene for Diamond-Blackfan anemia. Haematologica. 2017 Mar; 102(3):e93-e96. doi: 10.3324/haematol.2016.153932. [download]
- Ikeda Y, Kiyotani K, Yew PY, Sato S, Imai Y, Yamaguchi R, Miyano S, Fujiwara K, Hasegawa K, Nakamura Y. Clinical significance of T cell clonality and expression levels of immune-related genes in endometrial cancer. Oncol Rep. 2017 May; 37(5):2603-2610. doi: 10.3892/or.2017.5536. [download]
- Kato I, Nishinaka Y, Nakamura M, Akarca AU, Niwa A, Ozawa H, Yoshida K, Mori M, Wang D, Morita M, Ueno H, Shiozawa Y, Shiraishi Y, Miyano S, Gupta R, Umeda K, Watanabe K, Koh K, Adachi S, Heike T, Saito MK, Sanada M, Ogawa S, Marafioti T, Watanabe A, Nakahata T, Enver T. Hypoxic adaptation of leukemic cells infiltrating the CNS affords a therapeutic strategy targeting VEGFA. Blood. 2017 Jun 8; 129(23):3126-3129. doi: 10.1182/Blood-2016-06-721712. [download]
- Kato M, Ishimaru S, Seki M, Yoshida K, Shiraishi Y, Chiba K, Kakiuchi N, Sato Y, Ueno H, Tanaka H, Inukai T, Tomizawa D, Hasegawa D, Osumi T, Arakawa Y, Aoki T, Okuya M, Kaizu K, Kato K, Taneyama Y, Goto H, Taki T, Takagi M, Sanada M, Koh K, Takita J, Miyano S, Ogawa S, Ohara A, Tsuchida M, Manabe A. Long-term outcome of 6-month maintenance chemotherapy for acute lymphoblastic leukemia in children. Leukemia. 2017 Mar; 31(3):580-584. doi: 10.1038/leu.2016.274. [download]
- Lyons E, Sheridan P, Tremmel G, Miyano S, Sugano S. Large-scale DNA Barcode Library Generation for Biomolecule Identification in High-throughput Screens. Sci Rep. 2017 Oct 24;7(1):13899. doi: 10.1038/s41598-017-12825-2. [download]
- Makishima H, Yoshizato T, Yoshida K, Sekeres MA, Radivoyevitch T, Suzuki H, Przychodzen B, Nagata Y, Meggendorfer M, Sanada M, Okuno Y, Hirsch C, Kuzmanovic T, Sato Y, Sato-Otsubo A, LaFramboise T, Hosono N, Shiraishi Y, Chiba K, Haferlach C, Kern W, Tanaka H, Shiozawa Y, Gómez-Seguí I, Husseinzadeh HD, Thota S, Guinta KM, Dienes B, Nakamaki T, Miyawaki S, Saunthararajah Y, Chiba S, Miyano S, Shih LY, Haferlach T, Ogawa S, Maciejewski JP. Dynamics of clonal evolution in myelodysplastic syndromes. Nat Genet. 2017 Feb; 49(2):204-212. doi: 10.1038/ng.3742. [download]
- Matsui Y, Niida A, Uchi R, Mimori K, Miyano S, Shimamura T. phyC: Clustering cancer evolutionary trees. PLoS Comput Biol. 2017 May 1; 13(5):e1005509. doi: 10.1371/journal.pcbi.1005509. [download]
- Mitani K, Nagata Y, Sasaki K, Yoshida K, Chiba K, Tanaka H, Shiraishi Y, Miyano S, Makishima H, Nakamura Y, Nakamura Y, Ichikawa M, Ogawa S. Somatic mosaicism in chronic myeloid leukemia in remission. Blood. 2016 Dec 15;128(24):2863-2866. doi: 10.1182/Blood-2016-06-723494. [download]
- Miyamoto T, Tanikawa C, Yodsurang V, Zhang YZ, Imoto S, Yamaguchi R, Miyano S, Nakagawa H, Matsuda K. Identification of a p53-repressed gene module in breast cancer cells. Oncotarget. 2017 Jul 26; 8(34):55821-55836. doi: 10.18632/oncotarget.19608. [download]
- Morita T, Rahman A, Hasegawa T, Ozaki A, Tanimoto T. The potential possibility of symptom checker tools. Int J Health Policy Manag. 2017 Oct 1;6:615-616. doi: 10.15171/ijhpm.2017.41. PMID: 28949479; PMCID: PMC5627791.
- Moriyama T, Shiraishi Y, Chiba K, Yamaguchi R, Imoto S, Miyano S. OVarCall: Bayesian Mutation Calling Method Utilizing Overlapping Paired-End Reads. IEEE Trans Nanobioscience. 2017 Mar; 16(2):116-122. doi: 10.1109/TNB.2017.2670601. [download]
- Muramatsu H, Okuno Y, Yoshida K, Shiraishi Y, doi:saki S, Narita A, Sakaguchi H, Kawashima N, Wang X, Xu Y, Chiba K, Tanaka H, Hama A, Sanada M, Takahashi Y, Kanno H, Yamaguchi H, Ohga S, Manabe A, Harigae H, Kunishima S, Ishii E, Kobayashi M, Koike K, Watanabe K, Ito E, Takata M, Yabe M, Ogawa S, Miyano S, Kojima S. Clinical utility of next-generation sequencing for inherited bone marrow failure syndromes. Genet Med. 2017 Jul; 19(7):796-802. doi: 10.1038/gim.2016.197. [download]
- Nagata H, Kozaki KI, Muramatsu T, Hiramoto H, Tanimoto K, Fujiwara N, Imoto S, Ichikawa D, Otsuji E, Miyano S, Kawano T, Inazawa J. Genome-wide screening of DNA methylation associated with lymph node metastasis in esophageal squamous cell carcinoma. Oncotarget. 2017 Jun 6; 8(23):37740-37750. doi: 10.18632/oncotarget.17147. [download]
- Nguyen TB, Sakata-Yanagimoto M, Asabe Y, Matsubara D, Kano J, Yoshida K, Shiraishi Y, Chiba K, Tanaka H, Miyano S, Izutsu K, Nakamura N, Takeuchi K, Miyoshi H, Ohshima K, Minowa T, Ogawa S, Noguchi M, Chiba S. Identification of cell-type-specific mutations in nodal T-cell lymphomas. Blood Cancer J. 2017 Jan 6; 7(1):e516. doi: 10.1038/bcj.2016.122. [download]
- Okada A, Chiba K, Tanaka H, Miyano S, Shiraishi Y. A framework for generating interactive reports for cancer genome analysis. J Open Source Softw. 2017 Dec 6; 2(20): 457. doi: 10.21105/joss.00457. [download]
- Park H, Niida A, Imoto S, Miyano S. Interaction-Based Feature Selection for Uncovering Cancer Driver Genes Through Copy Number-Driven Expression Level. J Comput Biol. 2017 Feb; 24(2):138-152. doi: 10.1089/cmb.2016.0140. [download]
- Park H, Shiraishi Y, Imoto S, Miyano S. A Novel Adaptive Penalized Logistic Regression for Uncovering Biomarker Associated with Anti-Cancer Drug Sensitivity. IEEE/ACM Trans Comput Biol Bioinform. 2017 Jul-Aug; 14(4):771-782. doi: 10.1109/TCBB.2016.2561937. [download]
- Sato R, Shibata T, Tanaka Y, Kato C, Yamaguchi K, Furukawa Y, Shimizu E, Yamaguchi R, Imoto S, Miyano S, Miyake K. Requirement of glycosylation machinery in TLR responses revealed by CRISPR/Cas9 screening. Int Immunol. 2017 Aug 1;29(8):347-355. doi: 10.1093/intimm/dxx044. [download]
- Seki M, Kimura S, Isobe T, Yoshida K, Ueno H, Nakajima-Takagi Y, Wang C, Lin L, Kon A, Suzuki H, Shiozawa Y, Kataoka K, Fujii Y, Shiraishi Y, Chiba K, Tanaka H, Shimamura T, Masuda K, Kawamoto H, Ohki K, Kato M, Arakawa Y, Koh K, Hanada R, Moritake H, Akiyama M, Kobayashi R, Deguchi T, Hashii Y, Imamura T, Sato A, Kiyokawa N, Oka A, Hayashi Y, Takagi M, Manabe A, Ohara A, Horibe K, Sanada M, Iwama A, Mano H, Miyano S, Ogawa S, Takita J. Recurrent SPI1 (PU.1) fusions in high-risk pediatric T cell acute lymphoblastic leukemia. Nat Genet. 2017 Aug; 49(8):1274-1281. doi: 10.1038/ng.3900. [download]
- Sekinaka Y, Mitsuiki N, Imai K, Yabe M, Yabe H, Mitsui-Sekinaka K, Honma K, Takagi M, Arai A, Yoshida K, Okuno Y, Shiraishi Y, Chiba K, Tanaka H, Miyano S, Muramatsu H, Kojima S, Hira A, Takata M, Ohara O, Ogawa S, Morio T, Nonoyama S. Common Variable Immunodeficiency Caused by FANC Mutations. J Clin Immunol. 2017 Jul; 37(5):434-444. doi: 10.1007/s10875-017-0396-4. [download]
- Shiba N, Yoshida K, Shiraishi Y, Okuno Y, Yamato G, Hara Y, Nagata Y, Chiba K, Tanaka H, Terui K, Kato M, Park MJ, Ohki K, Shimada A, Takita J, Tomizawa D, Kudo K, Arakawa H, Adachi S, Taga T, Tawa A, Ito E, Horibe K, Sanada M, Miyano S, Ogawa S, Hayashi Y. Whole-exome sequencing reveals the spectrum of gene mutations and the clonal evolution patterns in paediatric acute myeloid leukaemia. Br J Haematol. 2016 Nov; 175(3):476-489. doi: 10.1111/bjh.14247. [download]
- Shiozawa Y, Malcovati L, Gallì A, Pellagatti A, Karimi M, Sato-Otsubo A, Sato Y, Suzuki H, Yoshizato T, Yoshida K, Shiraishi Y, Chiba K, Makishima H, Boultwood J, Hellström-Lindberg E, Miyano S, Cazzola M, Ogawa S. Gene expression and risk of leukemic transformation in myelodysplasia. Blood. 2017 Dec 14; 130(24):2642-2653. doi: 10.1182/Blood-2017-05-783050. Erratum in: Blood. 2018 Aug 23;132(8):869-875. doi: 10.1182/blood-2018-07-863134. [download]
- Sun QY, Ding LW, Tan KT, Chien W, Mayakonda A, Lin DC, Loh XY, Xiao JF, Meggendorfer M, Alpermann T, Garg M, Lim SL, Madan V, Hattori N, Nagata Y, Miyano S, Yeoh AE, Hou HA, Jiang YY, Takao S, Liu LZ, Tan SZ, Lill M, Hayashi M, Kinoshita A, Kantarjian HM, Kornblau SM, Ogawa S, Haferlach T, Yang H, Koeffler HP. Ordering of mutations in acute myeloid leukemia with partial tandem duplication of MLL (MLL-PTD). Leukemia. 2017 Jan; 31(1):1-10. doi: 10.1038/leu.2016.160. [download]
- Takagi M, Ogata S, Ueno H, Yoshida K, Yeh T, Hoshino A, Piao J, Yamashita M, Nanya M, Okano T, Kajiwara M, Kanegane H, Muramatsu H, Okuno Y, Shiraishi Y, Chiba K, Tanaka H, Bando Y, Kato M, Hayashi Y, Miyano S, Imai K, Ogawa S, Kojima S, Morio T. Haploinsufficiency of TNFAIP3 (A20) by germline mutation is involved in autoimmune lymphoproliferative syndrome. J Allergy Clin Immunol. 2017 Jun; 139(6):1914-1922. doi: 10.1016/j.jaci.2016.09.038. [download]
- Takagi M, Yoshida M, Nemoto Y, Tamaichi H, Tsuchida R, Seki M, Uryu K, Nishii R, Miyamoto S, Saito M, Hanada R, Kaneko H, Miyano S, Kataoka K, Yoshida K, Ohira M, Hayashi Y, Nakagawara A, Ogawa S, Mizutani S, Takita J. Loss of DNA Damage Response in Neuroblastoma and Utility of a PARP Inhibitor. J Natl Cancer Inst. 2017 Nov 1; 109(11):djx062. doi: 10.1093/jnci/djx062. [download]
- Takahashi Y, Sugimachi K, Yamamoto K, Niida A, Shimamura T, Sato T, Watanabe M, Tanaka J, Kudo S, Sugihara K, Hase K, Kusunoki M, Yamada K, Shimada Y, Moriya Y, Suzuki Y, Miyano S, Mori M, Mimori K. Japanese genome-wide association study identifies a significant colorectal cancer susceptibility locus at chromosome 10p14. Cancer Sci. 2017 Nov;108(11):2239-2247. doi: 10.1111/cas.13391. [download]
- Takano Y, Masuda T, Iinuma H, Yamaguchi R, Sato K, Tobo T, Hirata H, Kuroda Y, Nambara S, Hayashi N, Iguchi T, Ito S, Eguchi H, Ochiya T, Yanaga K, Miyano S, Mimori K. Circulating exosomal microRNA-203 is associated with metastasis possibly via inducing tumor-associated macrophages in colorectal cancer. Oncotarget. 2017 Aug 7;8(45):78598-78613. doi: 10.18632/Oncotarget.20009. [download]
- Tanikawa C, Zhang YZ, Yamamoto R, Tsuda Y, Tanaka M, Funauchi Y, Mori J, Imoto S, Yamaguchi R, Nakamura Y, Miyano S, Nakagawa H, Matsuda K. The Transcriptional Landscape of p53 Signalling Pathway. EBioMedicine. 2017 Jun; 20:109-119. doi: 10.1016/j.ebiom.2017.05.017. [download]
- Togasaki E, Takeda J, Yoshida K, Shiozawa Y, Takeuchi M, Oshima M, Saraya A, Iwama A, Yokote K, Sakaida E, Hirase C, Takeshita A, Imai K, Okumura H, Morishita Y, Usui N, Takahashi N, Fujisawa S, Shiraishi Y, Chiba K, Tanaka H, Kiyoi H, Ohnishi K, Ohtake S, Asou N, Kobayashi Y, Miyazaki Y, Miyano S, Ogawa S, Matsumura I, Nakaseko C, Naoe T. Frequent somatic mutations in epigenetic regulators in newly diagnosed chronic myeloid leukemia. Blood Cancer J. 2017 Apr 28; 7(4):e559. doi: 10.1038/bcj.2017.36. [download]
- Tominaga K, Shimamura T, Kimura N, Murayama T, Matsubara D, Kanauchi H, Niida A, Shimizu S, Nishioka K, Tsuji EI, Yano M, Sugano S, Shimono Y, Ishii H, Saya H, Mori M, Akashi K, Tada KI, Ogawa T, Tojo A, Miyano S, Gotoh N. Addiction to the IGF2-ID1-IGF2 circuit for maintenance of the breast cancer stem-like cells. Oncogene. 2017 Mar 2;36(9):1276-1286. doi: 10.1038/onc.2016.293. [download]
- Tsuda Y, Tanikawa C, Miyamoto T, Hirata M, Yodsurang V, Zhang YZ, Imoto S, Yamaguchi R, Miyano S, Takayanagi H, Kawano H, Nakagawa H, Tanaka S, Matsuda K. Identification of a p53 target, CD137L, that mediates growth suppression and immune response of osteosarcoma cells. Sci Rep. 2017 Sep 6; 7(1):10739. doi: 10.1038/s41598-017-11208-x. [download]
- Uryu K, Nishimura R, Kataoka K, Sato Y, Nakazawa A, Suzuki H, Yoshida K, Seki M, Hiwatari M, Isobe T, Shiraishi Y, Chiba K, Tanaka H, Miyano S, Koh K, Hanada R, Oka A, Hayashi Y, Ohira M, Kamijo T, Nagase H, Takimoto T, Tajiri T, Nakagawara A, Ogawa S, Takita J. Identification of the genetic and clinical characteristics of neuroblastomas using genome-wide analysis. Oncotarget. 2017 Nov 18;8(64):107513-107529. doi: 10.18632/oncotarget.22495. [download]
- Yamato G, Shiba N, Yoshida K, Shiraishi Y, Hara Y, Ohki K, Okubo J, Okuno H, Chiba K, Tanaka H, Kinoshita A, Moritake H, Kiyokawa N, Tomizawa D, Park MJ, Sotomatsu M, Taga T, Adachi S, Tawa A, Horibe K, Arakawa H, Miyano S, Ogawa S, Hayashi Y. ASXL2 mutations are frequently found in pediatric AML patients with t(8;21)/ RUNX1-RUNX1T1 and associated with a better prognosis. Genes Chromosomes Cancer. 2017 May; 56(5):382-393. doi: 10.1002/gcc.22443. [download]
- Yoshizato T, Nannya Y, Atsuta Y, Shiozawa Y, Iijima-Yamashita Y, Yoshida K, Shiraishi Y, Suzuki H, Nagata Y, Sato Y, Kakiuchi N, Matsuo K, Onizuka M, Kataoka K, Chiba K, Tanaka H, Ueno H, Nakagawa MM, Przychodzen B, Haferlach C, Kern W, Aoki K, Itonaga H, Kanda Y, Sekeres MA, Maciejewski JP, Haferlach T, Miyazaki Y, Horibe K, Sanada M, Miyano S, Makishima H, Ogawa S. Genetic abnormalities in myelodysplasia and secondary acute myeloid leukemia: impact on outcome of stem cell transplantation. Blood. 2017 Apr 27; 129(17):2347-2358. doi: 10.1182/Blood-2016-12-754796. [download]
- Zhang YZ, Imoto S, Miyano S, Yamaguchi R. Reconstruction of high read-depth signals from low-depth whole genome sequencing data using deep learning. 2017 IEEE International Conference on Bioinformatics and Biomedicine (BIBM). 2017 Nov; 1227-1232. doi: 10.1109/BIBM.2017.8217832. [download]
- Zhang YZ, Yamaguchi R, Imoto S, Miyano S. Sequence-specific bias correction for RNA-seq data using recurrent neural networks. BMC Genomics. 2017 Jan 25; 18(Suppl 1):1044. doi: 10.1186/s12864-016-3262-5. [download]
- Mori T, Nagata Y, Makishima H, Sanada M, Shiozawa Y, Kon A, Yoshizato T, Sato-Otsubo A, Kataoka K, Shiraishi Y, Chiba K, Tanaka H, Ishiyama K, Miyawaki S, Mori H, Nakamaki T, Kihara R, Kiyoi H, Koeffler HP, Shih LY, Miyano S, Naoe T, Haferlach C, Kern W, Haferlach T, Ogawa S, Yoshida K. Somatic PHF6 mutations in 1760 cases with various myeloid neoplasms. Leukemia. 2016 Nov; 30(11):2270-2273. doi: 10.1038/leu.2016.212. [download]
- Arashiki N, Takakuwa Y, Mohandas N, Hale J, Yoshida K, Ogura H, Utsugisawa T, Ohga S, Miyano S, Ogawa S, Kojima S, Kanno H. ATP11C is a major flippase in human erythrocytes and its defect causes congenital hemolytic anemia. Haematologica. 2016 May; 101(5):559-65. doi: 10.3324/haematol.2016.142273. [download]
- Ayada E, Hasegawa T, Niida A, Miyano S, Imoto S. Binary contingency table method for analysing gene mutation in cancer genome. Int. J Bioinform Res Appl. 2016 Aug 1; 12(3): 211-226. doi: 10.1504/IJBRA.2016.078231. [download]
- Chapman CG, Yamaguchi R, Tamura K, Weidner J, Imoto S, Kwon J, Fang H, Yew PY, Marino SR, Miyano S, Nakamura Y, Kiyotani K. Characterization of T-cell Receptor Repertoire in Inflamed Tissues of Patients with Crohn's Disease Through Deep Sequencing. Inflamm Bowel Dis. 2016 Jun; 22(6):1275-1285. doi: 10.1097/MIB.0000000000000752. [download]
- Fujimoto A, Furuta M, Totoki Y, Tsunoda T, Kato M, Shiraishi Y, Tanaka H, Taniguchi H, Kawakami Y, Ueno M, Gotoh K, Ariizumi S, Wardell CP, Hayami S, Nakamura T, Aikata H, Arihiro K, Boroevich KA, Abe T, Nakano K, Maejima K, Sasaki-Oku A, Ohsawa A, Shibuya T, Nakamura H, Hama N, Hosoda F, Arai Y, Ohashi S, Urushidate T, Nagae G, Yamamoto S, Ueda H, Tatsuno K, Ojima H, Hiraoka N, Okusaka T, Kubo M, Marubashi S, Yamada T, Hirano S, Yamamoto M, Ohdan H, Shimada K, Ishikawa O, Yamaue H, Chayama K, Miyano S, Aburatani H, Shibata T, Nakagawa H. Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nat Genet. 2016 May; 48(5):500-9. doi: 10.1038/ng.3547. Erratum in: Nat Genet. 2016 May 27; 48(6):700. doi: 10.1038/ng0616-700a. [download]
- Hasegawa T, Niida A, Mori T, Shimamura T, Yamaguchi R, Miyano S, Akutsu T, Imoto S. A likelihood-free filtering method via approximate Bayesian computation in evaluating biological simulation models. Comput Stat Data Anal. 2016 Feb; 94: 63-74. doi: https://doi.org/10.1016/j.csda.2015.08.003. [download]
- Hasegawa T, Kojima K, Kawai Y, Misawa K, Mimori T, Nagasaki M. AP-SKAT: a highly efficient genome-wide rare-variant association test. BMC Genomics. 2016 Sep 21;17:745. doi: 10.1186/s12864-016-3094-3. PMID: 27654840; PMCID: PMC5031335.
- Imashuku S, Muramatsu H, Sugihara T, Okuno Y, Wang X, Yoshida K, Kato A, Kato K, Tatsumi Y, Hattori A, Kita S, Oe K, Sueyoshi A, Usui T, Shiraishi Y, Chiba K, Tanaka H, Miyano S, Ogawa S, Kojima S, Kanno H. PIEZO1 gene mutation in a Japanese family with hereditary high phosphatidylcholine hemolytic anemia and hemochromatosis-induced diabetes mellitus. Int J Hematol. 2016 Jul; 104(1):125-9. doi: 10.1007/s12185-016-1970-x. [download]
- Kataoka K, Shiraishi Y, Takeda Y, Sakata S, Matsumoto M, Nagano S, Maeda T, Nagata Y, Kitanaka A, Mizuno S, Tanaka H, Chiba K, Ito S, Watatani Y, Kakiuchi N, Suzuki H, Yoshizato T, Yoshida K, Sanada M, Itonaga H, Imaizumi Y, Totoki Y, Munakata W, Nakamura H, Hama N, Shide K, Kubuki Y, Hidaka T, Kameda T, Masuda K, Minato N, Kashiwase K, Izutsu K, Takaori-Kondo A, Miyazaki Y, Takahashi S, Shibata T, Kawamoto H, Akatsuka Y, Shimoda K, Takeuchi K, Seya T, Miyano S, Ogawa S. Aberrant PD-L1 expression through 3'-UTR disruption in multiple cancers. Nature. 2016 Jun 16; 534(7607):402-6. doi: 10.1038/nature18294. [download]
- Kato M, Seki M, Yoshida K, Sato Y, Oyama R, Arakawa Y, Kishimoto H, Taki T, Akiyama M, Shiraishi Y, Chiba K, Tanaka H, Mitsuiki N, Kajiwara M, Mizutani S, Sanada M, Miyano S, Ogawa S, Koh K, Takita J. Genomic analysis of clonal origin of Langerhans cell histiocytosis following acute lymphoblastic leukaemia. Br J Haematol. 2016 Oct; 175(1): 169-72. doi: 10.1111/bjh.13841. [download]
- Ki Kim S, Ueda Y, Hatano E, Kakiuchi N, Takeda H, Goto T, Shimizu T, Yoshida K, Ikura Y, Shiraishi Y, Chiba K, Tanaka H, Miyano S, Uemoto S, Chiba T, Ogawa S, Marusawa H. TERT promoter mutations and chromosome 8p loss are characteristic of nonalcoholic fatty liver disease-related hepatocellular carcinoma. Int J Cancer. 2016 Dec 1; 139(11):2512-8. doi: 10.1002/ijc.30379. [download]
- Kitamura K, Okuno Y, Yoshida K, Sanada M, Shiraishi Y, Muramatsu H, Kobayashi R, Furukawa K, Miyano S, Kojima S, Ogawa S, Kunishima S. Functional characterization of a novel GFI1B mutation causing congenital macrothrombocytopenia. J Thromb Haemost. 2016 Jul;14(7):1462-1469. doi: 10.1111/jth.13350. [download]
- Koso H, Yi H, Sheridan P, Miyano S, Ino Y, Todo T, Watanabe S. Identification of RNA-Binding Protein LARP4B as a Tumor Suppressor in Glioma. Cancer Res. 2016 Apr 15; 76(8):2254-2264. doi: 10.1158/0008-5472.CAN-15-2308. [download]
- Kurashige J, Hasegawa T, Niida A, Sugimachi K, Deng N, Mima K, Uchi R, Sawada G, Takahashi Y, Eguchi H, Inomata M, Kitano S, Fukagawa T, Sasako M, Sasaki H, Sasaki S, Mori M, Yanagihara K, Baba H, Miyano S, Tan P, Mimori K. Integrated Molecular Profiling of Human Gastric Cancer Identifies DDR2 as a Potential Regulator of Peritoneal Dissemination. Sci Rep. 2016 Mar 3; 6:22371. doi: 10.1038/srep22371. [download]
- Madan V, Shyamsunder P, Han L, Mayakonda A, Nagata Y, Sundaresan J, Kanojia D, Yoshida K, Ganesan S, Hattori N, Fulton N, Tan KT, Alpermann T, Kuo MC, Rostami S, Matthews J, Sanada M, Liu LZ, Shiraishi Y, Miyano S, Chendamarai E, Hou HA, Malnassy G, Ma T, Garg M, Ding LW, Sun QY, Chien W, Ikezoe T, Lill M, Biondi A, Larson RA, Powell BL, Lübbert M, Chng WJ, Tien HF, Heuser M, Ganser A, Koren-Michowitz M, Kornblau SM, Kantarjian HM, Nowak D, Hofmann WK, Yang H, Stock W, Ghavamzadeh A, Alimoghaddam K, Haferlach T, Ogawa S, Shih LY, Mathews V, Koeffler HP. Comprehensive mutational analysis of primary and relapse acute promyelocytic leukemia. Leukemia. 2016 Aug; 30(8):1672-1681. doi: 10.1038/leu.2016.69. Erratum in: Leukemia. 2016 Dec; 30(12 ):2430. doi: 10.1038/leu.2016.237. [download]
- Mao Y, Tamura T, Yuki Y, Abe D, Tamada Y, Imoto S, Tanaka H, Homma H, Tagawa K, Miyano S, Okazawa H. The hnRNP-Htt axis regulates necrotic cell death induced by transcriptional repression through impaired RNA splicing. Cell Death Dis. 2016 Apr 28; 7(4):e2207. doi: 10.1038/cddis.2016.101. [download]
- Kojima K, Kawai Y, Nariai N, Mimori T, Hasegawa T, Nagasaki M. Short tandem repeat number estimation from paired-end reads for multiple individuals by considering a coalescent tree. BMC Genomics. 2016 Aug 31;17(Suppl 5):494. doi: 10.1186/s12864-016-2821-0. PMID: 27586631; PMCID: PMC5009668.
- Koshiba S, Motoike I, Kojima K, Hasegawa T, Shirota M, Saito T, Saigusa D, Danjoh I, Katsuoka F, Ogishima S, Kawai Y, Yamaguchi-Kabata Y, Sakurai M, Hirano S, Nakata J, Motohashi H, Hozawa A, Kuriyama S, Minegishi N, Nagasaki M, Takai-Igarashi T, Fuse N, Kiyomoto H, Sugawara J, Suzuki Y, Kure S, Yaegashi N, Tanabe O, Kinoshita K, Yasuda J, Yamamoto M. The structural origin of metabolic quantitative diversity. Sci Rep. 2016 Aug 16;6:31463. doi: 10.1038/srep31463. PMID: 27528366; PMCID: PMC4985752
- Matsui Y, Mizuta M, Ito S, Miyano S, Shimamura T. D3M: detection of differential distributions of methylation levels. Bioinformatics. 2016 Aug 1;32(15):2248-2255. doi: 10.1093/Bioinformatics/btw138. [download]
- Muramatsu T, Kozaki KI, Imoto S, Yamaguchi R, Tsuda H, Kawano T, Fujiwara N, Morishita M, Miyano S, Inazawa J. The hypusine cascade promotes cancer progression and metastasis through the regulation of RhoA in squamous cell carcinoma. Oncogene. 2016 Oct 6; 35(40):5304-5316. doi: 10.1038/onc.2016.71. [download]
- Nagata Y, Kontani K, Enami T, Kataoka K, Ishii R, Totoki Y, Kataoka TR, Hirata M, Aoki K, Nakano K, Kitanaka A, Sakata-Yanagimoto M, Egami S, Shiraishi Y, Chiba K, Tanaka H, Shiozawa Y, Yoshizato T, Suzuki H, Kon A, Yoshida K, Sato Y, Sato-Otsubo A, Sanada M, Munakata W, Nakamura H, Hama N, Miyano S, Nureki O, Shibata T, Haga H, Shimoda K, Katada T, Chiba S, Watanabe T, Ogawa S. Variegated RHOA mutations in adult T-cell leukemia/lymphoma. Blood. 2016 Feb 4;127(5):596-604. doi: 10.1182/Blood-2015-06-644948. [download]
- Sakurai M, Kasahara H, Yoshida K, Yoshimi A, Kunimoto H, Watanabe N, Shiraishi Y, Chiba K, Tanaka H, Harada Y, Harada H, Kawakita T, Kurokawa M, Miyano S, Takahashi S, Ogawa S, Okamoto S, Nakajima H. Genetic basis of myeloid transformation in familial platelet disorder/acute myeloid leukemia patients with haploinsufficient RUNX1 allele. Blood Cancer J. 2016 Feb 5; 6(2): e392. doi: 10.1038/bcj.2015.81. [download]
- Sawada G, Niida A, Uchi R, Hirata H, Shimamura T, Suzuki Y, Shiraishi Y, Chiba K, Imoto S, Takahashi Y, Iwaya T, Sudo T, Hayashi T, Takai H, Kawasaki Y, Matsukawa T, Eguchi H, Sugimachi K, Tanaka F, Suzuki H, Yamamoto K, Ishii H, Shimizu M, Yamazaki H, Yamazaki M, Tachimori Y, Kajiyama Y, Natsugoe S, Fujita H, Mafune K, Tanaka Y, Kelsell DP, Scott CA, Tsuji S, Yachida S, Shibata T, Sugano S, Doki Y, Akiyama T, Aburatani H, Ogawa S, Miyano S, Mori M, Mimori K. Genomic Landscape of Esophageal Squamous Cell Carcinoma in a Japanese Population. Gastroenterology. 2016 May; 150(5):1171-1182. doi: 10.1053/j.gastro.2016.01.035. [download]
- Sugimachi K, Yamaguchi R, Eguchi H, Ueda M, Niida A, Sakimura S, Hirata H, Uchi R, Shinden Y, Iguchi T, Morita K, Yamamoto K, Miyano S, Mori M, Maehara Y, Mimori K. 8q24 Polymorphisms and Diabetes Mellitus Regulate Apolipoprotein A-IV in Colorectal Carcinogenesis. Ann Surg Oncol. 2016 Jul 07; 23(Suppl 4):546-551. doi: 10.1245/s10434-016-5374-1. [download]
- Tamura K, Hazama S, Yamaguchi R, Imoto S, Takenouchi H, Inoue Y, Kanekiyo S, Shindo Y, Miyano S, Nakamura Y, Kiyotani K. Characterization of the T Cell Repertoire by deep T cell receptor sequencing in tissues and blood from patients with advanced colorectal cancer. Oncol Lett. 2016 Jun; 11(6):3643-3649. doi: 10.3892/ol.2016.4465. [download]
- Tsujita Y, Mitsui-Sekinaka K, Imai K, Yeh TW, Mitsuiki N, Asano T, Ohnishi H, Kato Z, Sekinaka Y, Zaha K, Kato T, Okano T, Takashima T, Kobayashi K, Kimura M, Kunitsu T, Maruo Y, Kanegane H, Takagi M, Yoshida K, Okuno Y, Muramatsu H, Shiraishi Y, Chiba K, Tanaka H, Miyano S, Kojima S, Ogawa S, Ohara O, Okada S, Kobayashi M, Morio T, Nonoyama S. Phosphatase and tensin homolog (PTEN) mutation can cause activated phosphatidylinositol 3-kinase δ syndrome-like immunodeficiency. J Allergy Clin Immunol. 2016 Dec; 138(6):1672-1680.e10. doi: 10.1016/j.jaci.2016.03.055. [download]
- Uchi R, Takahashi Y, Niida A, Shimamura T, Hirata H, Sugimachi K, Sawada G, Iwaya T, Kurashige J, Shinden Y, Iguchi T, Eguchi H, Chiba K, Shiraishi Y, Nagae G, Yoshida K, Nagata Y, Haeno H, Yamamoto H, Ishii H, Doki Y, Iinuma H, Sasaki S, Nagayama S, Yamada K, Yachida S, Kato M, Shibata T, Oki E, Saeki H, Shirabe K, Oda Y, Maehara Y, Komune S, Mori M, Suzuki Y, Yamamoto K, Aburatani H, Ogawa S, Miyano S, Mimori K. Integrated Multiregional Analysis Proposing a New Model of Colorectal Cancer Evolution. PLoS Genet. 2016 Feb 18; 12(2):e1005778. doi: 10.1371/journal.pgen.1005778. Erratum in: PLoS Genet. 2017 May 19; 13(5):e1006798. doi: 10.1371/journal.pgen.1006798. [download]
- Ueda M, Iguchi T, Masuda T, Nakahara Y, Hirata H, Uchi R, Niida A, Momose K, Sakimura S, Chiba K, Eguchi H, Ito S, Sugimachi K, Yamasaki M, Suzuki Y, Miyano S, Doki Y, Mori M, Mimori K. Somatic mutations in plasma cell-free DNA are diagnostic markers for esophageal squamous cell carcinoma recurrence. Oncotarget. 2016 Sep 20;7(38):62280-62291. doi: 10.18632/Oncotarget.11409. [download]
- Yamaguchi K, Nagayama S, Shimizu E, Komura M, Yamaguchi R, Shibuya T, Arai M, Hatakeyama S, Ikenoue T, Ueno M, Miyano S, Imoto S, Furukawa Y. Reduced expression of APC-1B but not APC-1A by the deletion of promoter 1B is responsible for familial adenomatous polyposis. Sci Rep. 2016 May 24; 6:26011. doi: 10.1038/srep26011. [download]
- Yoshino T, Katayama K, Horiba Y, Munakata K, Yamaguchi R, Imoto S, Miyano S, Mima H, Watanabe K. Predicting Japanese Kampo formulas by analyzing database of medical records: a preliminary observational study. BMC Med Inform Decis Mak. 2016 Sep 13; 16(1):118. doi: 10.1186/s12911-016-0361-9. [download]
- Yoshino T, Katayama K, Horiba Y, Munakata K, Yamaguchi R, Imoto S, Miyano S, Mima H, Watanabe K, Mimura M. The Difference between the Two Representative Kampo Formulas for Treating Dysmenorrhea: An Observational Study. Evid Based Complement Alternat Med. 2016 Feb 24; 2016:3159617. doi: 10.1155/2016/3159617. [download]
- Chiba K, Shiraishi Y, Nagata Y, Yoshida K, Imoto S, Ogawa S, Miyano S. Genomon ITDetector: a tool for somatic internal tandem duplication detection from cancer genome sequencing data. Bioinformatics. 2015 Jan 1; 31(1):116-118. doi: 10.1093/Bioinformatics/btu593. [download]
- Fang H, Yamaguchi R, Liu X, Daigo Y, Yew PY, Tanikawa C, Matsuda K, Imoto S, Miyano S, Nakamura Y. Quantitative T cell repertoire analysis by deep cDNA sequencing of T cell receptor α and β chains using next-generation sequencing (NGS). Oncoimmunology. 2015 Jan 7; 3(12):e968467. doi: 10.4161/21624011.2014.968467. [download]
- Fujimoto A, Furuta M, Shiraishi Y, Gotoh K, Kawakami Y, Arihiro K, Nakamura T, Ueno M, Ariizumi S, Nguyen HH, Shigemizu D, Abe T, Boroevich KA, Nakano K, Sasaki A, Kitada R, Maejima K, Yamamoto Y, Tanaka H, Shibuya T, Shibata T, Ojima H, Shimada K, Hayami S, Shigekawa Y, Aikata H, Ohdan H, Marubashi S, Yamada T, Kubo M, Hirano S, Ishikawa O, Yamamoto M, Yamaue H, Chayama K, Miyano S, Tsunoda T, Nakagawa H. Whole-genome mutational landscape of liver cancers displaying biliary phenotype reveals hepatitis impact and molecular diversity. Nat Commun. 2015 Jan 30; 6:6120. doi: 10.1038/ncomms7120. [download]
- Garg M, Nagata Y, Kanojia D, Mayakonda A, Yoshida K, Haridas Keloth S, Zang ZJ, Okuno Y, Shiraishi Y, Chiba K, Tanaka H, Miyano S, Ding LW, Alpermann T, Sun QY, Lin DC, Chien W, Madan V, Liu LZ, Tan KT, Sampath A, Venkatesan S, Inokuchi K, Wakita S, Yamaguchi H, Chng WJ, Kham SK, Yeoh AE, Sanada M, Schiller J, Kreuzer KA, Kornblau SM, Kantarjian HM, Haferlach T, Lill M, Kuo MC, Shih LY, Blau IW, Blau O, Yang H, Ogawa S, Koeffler HP. Profiling of somatic mutations in acute myeloid leukemia with FLT3-ITD at diagnosis and relapse. Blood. 2015 Nov 26; 126(22):2491-501. doi: 10.1182/Blood-2015-05-646240. [download]
- Hasegawa T, Mori T, Yamaguchi R, Shimamura T, Miyano S, Imoto S, Akutsu T. Genomic data assimilation using a higher moment filtering technique for restoration of gene regulatory networks. BMC Syst Biol. 2015 Mar 13; 9:14. doi: 10.1186/s12918-015-0154-2. [download]
- Hira A, Yoshida K, Sato K, Okuno Y, Shiraishi Y, Chiba K, Tanaka H, Miyano S, Shimamoto A, Tahara H, Ito E, Kojima S, Kurumizaka H, Ogawa S, Takata M, Yabe H, Yabe M. Mutations in the gene encoding the E2 conjugating enzyme UBE2T cause Fanconi anemia. Am J Hum Genet. 2015 Jun 4; 96(6):1001-1007. doi: 10.1016/j.ajhg.2015.04.022. [download]
- Hoshino A, Nomura K, Hamashima T, Isobe T, Seki M, Hiwatari M, Yoshida K, Shiraishi Y, Chiba K, Tanaka H, Miyano S, Ogawa S, Takita J, Kanegane H. Aggressive transformation of anaplastic large cell lymphoma with increased number of ALK-translocated chromosomes. Int J Hematol. 2015 Feb; 101(2):198-202. doi: 10.1007/s12185-014-1701-0. [download]
- Huang D, Nagata Y, Grossmann V, Radivoyevitch T, Okuno Y, Nagae G, Hosono N, Schnittger S, Sanada M, Przychodzen B, Kon A, Polprasert C, Shen W, Clemente MJ, Phillips JG, Alpermann T, Yoshida K, Nadarajah N, Sekeres MA, Oakley K, Nguyen N, Shiraishi Y, Shiozawa Y, Chiba K, Tanaka H, Koeffler HP, Klein HU, Dugas M, Aburatani H, Miyano S, Haferlach C, Kern W, Haferlach T, Du Y, Ogawa S, Makishima H. BRCC3 mutations in myeloid neoplasms. Haematologica. 2015 Aug; 100(8):1051-1057. doi: 10.3324/haematol.2014.111989. [download]
- Ikenoue T, Yamaguchi K, Komura M, Imoto S, Yamaguchi R, Shimizu E, Kasuya S, Shibuya T, Hatakeyama S, Miyano S, Furukawa Y. Attenuated familial adenomatous polyposis with desmoids caused by an APC mutation. Hum Genome Var. 2015 Mar 26; 2:15011. doi: 10.1038/hgv.2015.11. [download]
- Ito S, Shiraishi Y, Shimamura T, Chiba K, Miyano S. High performance computing of a fusion gene detection pipeline on the K computer. 2015 IEEE International Conference on Bioinformatics and Biomedicine (BIBM). 2015 Nov; 1441-1447. doi: 10.1109/BIBM.2015.7359888. [download]
- Ito H, Shiwaku H, Yoshida C, Homma H, Luo H, Chen X, Fujita K, Musante L, Fischer U, Frints SG, Romano C, Ikeuchi Y, Shimamura T, Imoto S, Miyano S, Muramatsu SI, Kawauchi T, Hoshino M, Sudol M, Arumughan A, Wanker EE, Rich T, Schwartz C, Matsuzaki F, Bonni A, Kalscheuer VM, Okazawa H. In utero gene therapy rescues microcephaly caused by Pqbp1-hypofunction in neural stem progenitor cells. Mol Psychiatry. 2015 Apr; 20(4):459-471. doi: 10.1038/mp.2014.69. [download]
- Iwakawa R, Kohno T, Totoki Y, Shibata T, Tsuchihara K, Mimaki S, Tsuta K, Narita Y, Nishikawa R, Noguchi M, Harris CC, Robles AI, Yamaguchi R, Imoto S, Miyano S, Totsuka H, Yoshida T, Yokota J. Expression and clinical significance of genes frequently mutated in small cell lung cancers defined by whole exome/RNA sequencing. Carcinogenesis. 2015 Jun; 36(6):616-621. doi: 10.1093/carcin/bgv026. [download]
- Kanojia D, Nagata Y, Garg M, Lee DH, Sato A, Yoshida K, Sato Y, Sanada M, Mayakonda A, Bartenhagen C, Klein HU, Doan NB, Said JW, Mohith S, Gunasekar S, Shiraishi Y, Chiba K, Tanaka H, Miyano S, Myklebost O, Yang H, Dugas M, Meza-Zepeda LA, Silberman AW, Forscher C, Tyner JW, Ogawa S, Koeffler HP. Genomic landscape of liposarcoma. Oncotarget. 2015 Dec 15; 6(40):42429-4244. doi: 10.18632/oncotarget.6464. [download]
- Kataoka K, Nagata Y, Kitanaka A, Shiraishi Y, Shimamura T, Yasunaga J, Totoki Y, Chiba K, Sato-Otsubo A, Nagae G, Ishii R, Muto S, Kotani S, Watatani Y, Takeda J, Sanada M, Tanaka H, Suzuki H, Sato Y, Shiozawa Y, Yoshizato T, Yoshida K, Makishima H, Iwanaga M, Ma G, Nosaka K, Hishizawa M, Itonaga H, Imaizumi Y, Munakata W, Ogasawara H, Sato T, Sasai K, Muramoto K, Penova M, Kawaguchi T, Nakamura H, Hama N, Shide K, Kubuki Y, Hidaka T, Kameda T, Nakamaki T, Ishiyama K, Miyawaki S, Yoon SS, Tobinai K, Miyazaki Y, Takaori-Kondo A, Matsuda F, Takeuchi K, Nureki O, Aburatani H, Watanabe T, Shibata T, Matsuoka M, Miyano S, Shimoda K, Ogawa S. Integrated molecular analysis of adult T cell leukemia/lymphoma. Nat Genet. 2015 Nov; 47(11):1304-1315. doi: 10.1038/ng.3415. [download]
- Kawashima-Goto S, Imamura T, Seki M, Kato M, Yoshida K, Sugimoto A, Kaneda D, Fujiki A, Miyachi M, Nakatani T, Osone S, Ishida H, Taki T, Takita J, Shiraishi Y, Chiba K, Tanaka H, Miyano S, Ogawa S, Hosoi H. Identification of a homozygous JAK3 V674A mutation caused by acquired uniparental disomy in a relapsed early T-cell precursor ALL patient. Int J Hematol. 2015 Apr; 101(4):411-6. doi: 10.1007/s12185-014-1711-y. [download]
- Kayano M, Matsui H, Yamaguchi R, Imoto S, Miyano S. Gene set differential analysis of time course expression profiles via sparse estimation in functional logistic model with application to time-dependent biomarker detection. Biostatistics. 2016 Apr; 17(2):235-248. doi: 10.1093/biostatistics/kxv037. [download]
- Kurtovic-Kozaric A, Przychodzen B, Singh J, Konarska MM, Clemente MJ, Otrock ZK, Nakashima M, Hsi ED, Yoshida K, Shiraishi Y, Chiba K, Tanaka H, Miyano S, Ogawa S, Boultwood J, Makishima H, Maciejewski JP, Padgett RA. PRPF8 defects cause missplicing in myeloid malignancies. Leukemia. 2015 Jan; 29(1):126-136. doi: 10.1038/leu.2014.144. [download]
- Madan V, Kanojia D, Li J, Okamoto R, Sato-Otsubo A, Kohlmann A, Sanada M, Grossmann V, Sundaresan J, Shiraishi Y, Miyano S, Thol F, Ganser A, Yang H, Haferlach T, Ogawa S, Koeffler HP. Aberrant splicing of U12-type introns is the hallmark of ZRSR2 mutant myelodysplastic syndrome. Nat Commun. 2015 Jan 14; 6:6042. doi: 10.1038/ncomms7042. [download]
- Manolio TA, Abramowicz M, Al-Mulla F, Anderson W, Balling R, Berger AC, Bleyl S, Chakravarti A, Chantratita W, Chisholm RL, Dissanayake VH, Dunn M, Dzau VJ, Han BG, Hubbard T, Kolbe A, Korf B, Kubo M, Lasko P, Leego E, Mahasirimongkol S, Majumdar PP, Matthijs G, McLeod HL, Metspalu A, Meulien P, Miyano S, Naparstek Y, O'Rourke PP, Patrinos GP, Rehm HL, Relling MV, Rennert G, Rodriguez LL, Roden DM, Shuldiner AR, Sinha S, Tan P, Ulfendahl M, Ward R, Williams MS, Wong JE, Green ED, Ginsburg GS. Global implementation of genomic medicine: We are not alone. Sci Transl Med. 2015 Jun 3; 7(290):290ps13. doi: 10.1126/scitranslmed.aab0194. [download]
- Molenaar RJ, Thota S, Nagata Y, Patel B, Clemente M, Przychodzen B, Hirsh C, Viny AD, Hosano N, Bleeker FE, Meggendorfer M, Alpermann T, Shiraishi Y, Chiba K, Tanaka H, van Noorden CJ, Radivoyevitch T, Carraway HE, Makishima H, Miyano S, Sekeres MA, Ogawa S, Haferlach T, Maciejewski JP. Clinical and biological implications of ancestral and non-ancestral IDH1 and IDH2 mutations in myeloid neoplasms. Leukemia. 2015 Nov;29(11):2134-42. doi: 10.1038/leu.2015.91. [download]
- Nakata A, Yoshida R, Yamaguchi R, Yamauchi M, Tamada Y, Fujita A, Shimamura T, Imoto S, Higuchi T, Nomura M, Kimura T, Nokihara H, Higashiyama M, Kondoh K, Nishihara H, Tojo A, Yano S, Miyano S, Gotoh N. Elevated β-catenin pathway as a novel target for patients with resistance to EGF receptor targeting drugs. Sci Rep. 2015 Aug 13; 5:13076. doi: 10.1038/srep13076. [download]
- Okuno Y, Hoshino A, Muramatsu H, Kawashima N, Wang X, Yoshida K, Wada T, Gunji M, Toma T, Kato T, Shiraishi Y, Iwata A, Hori T, Kitoh T, Chiba K, Tanaka H, Sanada M, Takahashi Y, Nonoyama S, Ito M, Miyano S, Ogawa S, Kojima S, Kanegane H. Late-Onset Combined Immunodeficiency with a Novel IL2RG Mutation and Probable Revertant Somatic Mosaicism. J Clin Immunol. 2015 Oct; 35(7):610-614. doi: 10.1007/s10875-015-0202-0. [download]
- Ono A, Fujimoto A, Yamamoto Y, Akamatsu S, Hiraga N, Imamura M, Kawaoka T, Tsuge M, Abe H, Hayes CN, Miki D, Furuta M, Tsunoda T, Miyano S, Kubo M, Aikata H, Ochi H, Kawakami YI, Arihiro K, Ohdan H, Nakagawa H, Chayama K. Circulating Tumor DNA Analysis for Liver Cancers and Its Usefulness as a Liquid Biopsy. Cell Mol Gastroenterol Hepatol. 2015 Jun 17; 1(5):516-534. doi: 10.1016/j.jcmgh.2015.06.009. [download]
- Park H, Imoto S, Miyano S. Recursive Random Lasso (RRLasso) for Identifying Anti-Cancer Drug Targets. PLoS One. 2015 Nov 6; 10(11):e0141869. doi: 10.1371/journal.pone.0141869. [download]
- Park H, Niida A, Miyano S, Imoto S. Sparse overlapping group lasso for integrative multi-omics analysis. J Comput Biol. 2015 Feb; 22(2):73-84. doi: 10.1089/cmb.2014.0197. [download]
- Polprasert C, Schulze I, Sekeres MA, Makishima H, Przychodzen B, Hosono N, Singh J, Padgett RA, Gu X, Phillips JG, Clemente M, Parker Y, Lindner D, Dienes B, Jankowsky E, Saunthararajah Y, Du Y, Oakley K, Nguyen N, Mukherjee S, Pabst C, Godley LA, Churpek JE, Pollyea DA, Krug U, Berdel WE, Klein HU, Dugas M, Shiraishi Y, Chiba K, Tanaka H, Miyano S, Yoshida K, Ogawa S, Müller-Tidow C, Maciejewski JP. Inherited and Somatic Defects in DDX41 in Myeloid Neoplasms. Cancer Cell. 2015 May 11; 27(5):658-670. doi: 10.1016/j.ccell.2015.03.017. [download]
- Saini H, Raicar G, Sharma A, Lal S, Dehzangi A, Lyons J, Paliwal KK, Imoto S, Miyano S. Probabilistic expression of spatially varied amino acid dimers into general form of Chou׳s pseudo amino acid composition for protein fold recognition. J Theor Biol. 2015 Sep 7;380:291-8. doi: 10.1016/j.jtbi.2015.05.030. Epub 2015 Jun 12. PMID: 26079221 [download]
- Sakaguchi H, Muramatsu H, Okuno Y, Makishima H, Xu Y, Furukawa-Hibi Y, Wang X, Narita A, Yoshida K, Shiraishi Y, doi:saki S, Yoshida N, Hama A, Takahashi Y, Yamada K, Miyano S, Ogawa S, Maciejewski JP, Kojima S. Aberrant DNA Methylation Is Associated with a Poor Outcome in Juvenile Myelomonocytic Leukemia. PLoS One. 2015 Dec 31; 10(12):e0145394. doi: 10.1371/journal.pone.0145394. [download]
- Sawada G, Niida A, Hirata H, Komatsu H, Uchi R, Shimamura T, Takahashi Y, Kurashige J, Matsumura T, Ueo H, Takano Y, Ueda M, Sakimura S, Shinden Y, Eguchi H, Sudo T, Sugimachi K, Yamasaki M, Tanaka F, Tachimori Y, Kajiyama Y, Natsugoe S, Fujita H, Tanaka Y, Calin G, Miyano S, Doki Y, Mori M, Mimori K. An Integrative Analysis to Identify Driver Genes in Esophageal Squamous Cell Carcinoma. PLoS One. 2015 Oct 14; 10(10):e0139808. doi: 10.1371/journal.pone.0139808. [download]
- Seki M, Nishimura R, Yoshida K, Shimamura T, Shiraishi Y, Sato Y, Kato M, Chiba K, Tanaka H, Hoshino N, Nagae G, Shiozawa Y, Okuno Y, Hosoi H, Tanaka Y, Okita H, Miyachi M, Souzaki R, Taguchi T, Koh K, Hanada R, Kato K, Nomura Y, Akiyama M, Oka A, Igarashi T, Miyano S, Aburatani H, Hayashi Y, Ogawa S, Takita J. Integrated genetic and epigenetic analysis defines novel molecular subgroups in rhabdomyosarcoma. Nat Commun. 2015 Jul 3; 6:7557. doi: 10.1038/ncomms8557. [download]
- Shiota M, Yang X, Kubokawa M, Morishima T, Tanaka K, Mikami M, Yoshida K, Kikuchi M, Izawa K, Nishikomori R, Okuno Y, Wang X, Sakaguchi H, Muramatsu H, Kojima S, Miyano S, Ogawa S, Takagi M, Hata D, Kanegane H. Somatic mosaicism for a NRAS mutation associates with disparate clinical features in RAS-associated leukoproliferative disease: a report of two cases. J Clin Immunol. 2015 Jul; 35(5):454-8. doi: 10.1007/s10875-015-0163-3. [download]
- Shiraishi Y, Tremmel G, Miyano S, Stephens M. A Simple Model-Based Approach to Inferring and Visualizing Cancer Mutation Signatures. PLoS Genet. 2015 Dec 2; 11(12):e1005657. doi: 10.1371/journal.pgen.1005657. [download]
- Suzuki H, Aoki K, Chiba K, Sato Y, Shiozawa Y, Shiraishi Y, Shimamura T, Niida A, Motomura K, Ohka F, Yamamoto T, Tanahashi K, Ranjit M, Wakabayashi T, Yoshizato T, Kataoka K, Yoshida K, Nagata Y, Sato-Otsubo A, Tanaka H, Sanada M, Kondo Y, Nakamura H, Mizoguchi M, Abe T, Muragaki Y, Watanabe R, Ito I, Miyano S, Natsume A, Ogawa S. Mutational landscape and clonal architecture in grade II and III gliomas. Nat Genet. 2015 May;47(5):458-468. doi: 10.1038/ng.3273. [download]
- Tagawa K, Homma H, Saito A, Fujita K, Chen X, Imoto S, Oka T, Ito H, Motoki K, Yoshida C, Hatsuta H, Murayama S, Iwatsubo T, Miyano S, Okazawa H. Comprehensive phosphoproteome analysis unravels the core signaling network that initiates the earliest synapse pathology in preclinical Alzheimer's disease brain. Hum Mol Genet. 2015 Jan 15;24(2):540-558. doi: 10.1093/hmg/ddu475. [download]
- Takahashi Y, Sheridan P, Niida A, Sawada G, Uchi R, Mizuno H, Kurashige J, Sugimachi K, Sasaki S, Shimada Y, Hase K, Kusunoki M, Kudo S, Watanabe M, Yamada K, Sugihara K, Yamamoto H, Suzuki A, Doki Y, Miyano S, Mori M, Mimori K. The AURKA/TPX2 axis drives colon tumorigenesis cooperatively with MYC. Ann Oncol. 2015 May; 26(5):935-942. doi: 10.1093/annonc/mdv034. [download]
- Wang R, Yoshida K, Toki T, Sawada T, Uechi T, Okuno Y, Sato-Otsubo A, Kudo K, Kamimaki I, Kanezaki R, Shiraishi Y, Chiba K, Tanaka H, Terui K, Sato T, Iribe Y, Ohga S, Kuramitsu M, Hamaguchi I, Ohara A, Hara J, Goi K, Matsubara K, Koike K, Ishiguro A, Okamoto Y, Watanabe K, Kanno H, Kojima S, Miyano S, Kenmochi N, Ogawa S, Ito E. Loss of function mutations in RPL27 and RPS27 identified by whole-exome sequencing in Diamond-Blackfan anaemia. Br J Haematol. 2015 Mar;168(6):854-864. doi: 10.1111/bjh.13229. [download]
- Wang X, Muramatsu H, Okuno Y, Sakaguchi H, Yoshida K, Kawashima N, Xu Y, Shiraishi Y, Chiba K, Tanaka H, Saito S, Nakazawa Y, Masunari T, Hirose T, Elmahdi S, Narita A, doi:saki S, Ismael O, Makishima H, Hama A, Miyano S, Takahashi Y, Ogawa S, Kojima S. GATA2 and secondary mutations in familial myelodysplastic syndromes and pediatric myeloid malignancies. Haematologica. 2015 Oct;100(10):e398-401. doi: 10.3324/haematol.2015.127092. [download]
- Yamaguchi H, Sakaguchi H, Yoshida K, Yabe M, Yabe H, Okuno Y, Muramatsu H, Takahashi Y, Yui S, Shiraishi Y, Chiba K, Tanaka H, Miyano S, Inokuchi K, Ito E, Ogawa S, Kojima S. Clinical and genetic features of dyskeratosis congenita, cryptic dyskeratosis congenita, and Hoyeraal-Hreidarsson syndrome in Japan. Int J Hematol. 2015 Nov; 102(5):544-552. doi: 10.1007/s12185-015-1861-6. [download]
- Yamaguchi K, Komura M, Yamaguchi R, Imoto S, Shimizu E, Kasuya S, Shibuya T, Hatakeyama S, Takahashi N, Ikenoue T, Hata K, Tsurita G, Shinozaki M, Suzuki Y, Sugano S, Miyano S, Furukawa Y. Detection of APC mosaicism by next-generation sequencing in an FAP patient. J Hum Genet. 2015 May; 60(5):227-231. doi: 10.1038/jhg.2015.14. [download]
- Yamaguchi R, Imoto S, Miyano S. A TCR Sequence Data Analysis Pipeline: Tcrip. Immunopharmacogenomics. Nakamura, Y. (eds). Springer, Tokyo. 2015 Jan 01; 27-43. https://doi.org/10.1007/978-4-431-55726-5_2 [download]
- Yang X, Hoshino A, Taga T, Kunitsu T, Ikeda Y, Yasumi T, Yoshida K, Wada T, Miyake K, Kubota T, Okuno Y, Muramatsu H, Adachi Y, Miyano S, Ogawa S, Kojima S, Kanegane H. A female patient with incomplete hemophagocytic lymphohistiocytosis caused by a heterozygous XIAP mutation associated with non-random X-chromosome inactivation skewed towards the wild-type XIAP allele. J Clin Immunol. 2015 Apr; 35(3):244-8. doi: 10.1007/s10875-015-0144-6. [download]
- Yasuda T, Miyano S. Inferring the global structure of chromosomes from structural variations. BMC Genomics. 2015 Jan 21; 16 Suppl 2(Suppl 2):S13. doi: 10.1186/1471-2164-16-S2-S13. Erratum in: BMC Genomics. 2015 Apr 09; 16:276. doi: 10.1186/s12864-015-1338-2. [download]
- Yew PY, Alachkar H, Yamaguchi R, Kiyotani K, Fang H, Yap KL, Liu HT, Wickrema A, Artz A, van Besien K, Imoto S, Miyano S, Bishop MR, Stock W, Nakamura Y. Quantitative characterization of T-Cell Repertoire in allogeneic hematopoietic stem cell transplant recipients. Bone Marrow Transplant. 2015 Sep;50(9):1227-1234. doi: 10.1038/bmt.2015.133. [download]
- Yoshizato T, Dumitriu B, Hosokawda K, Makishima H, Yoshida K, Townsley D, Sato-Otsubo A, Sato Y, Liu D, Suzuki H, Wu CO, Shiraishi Y, Clemente MJ, Kataoka K, Shiozawa Y, Okuno Y, Chiba K, Tanaka H, Nagata Y, Katagiri T, Kon A, Sanada M, Scheinberg P, Miyano S, Maciejewski JP, Nakao S, Young NS, Ogawa S. Somatic Mutations and Clonal Hematopoiesis in Aplastic Anemia. N Engl J Med. 2015 Jul 2; 373(1):35-47. doi: 10.1056/NEJMoa1414799. [download]
- Fujita A, Miyano S. A tutorial to identify nonlinear associations in gene expression time series data. Methods Mol Biol. 2014 Jan 1; 1164:87-95. doi: 10.1007/978-1-4939-0805-9_8. Review [download]
- Shiraishi Y, Fujimoto A, Furuta M, Tanaka H, Chiba K, Boroevich KA, Abe T, Kawakami Y, Ueno M, Gotoh K, Ariizumi S, Shibuya T, Nakano K, Sasaki A, Maejima K, Kitada R, Hayami S, Shigekawa Y, Marubashi S, Yamada T, Kubo M, Ishikawa O, Aikata H, Arihiro K, Ohdan H, Yamamoto M, Yamaue H, Chayama K, Tsunoda T, Miyano S, Nakagawa H. Integrated analysis of whole genome and transcriptome sequencing reveals diverse transcriptomic aberrations driven by somatic genomic changes in liver cancers. PLoS One. 2014 Dec 19; 9(12):e114263. doi: 10.1371/journal.pone.0114263. [download]
- Arima C, Kajino T, Tamada Y, Imoto S, Shimada Y, Nakatochi M, Suzuki M, Isomura H, Yatabe Y, Yamaguchi T, Yanagisawa K, Miyano S, Takahashi T. Lung adenocarcinoma subtypes definable by lung development-related miRNA expression profiles in association with clinicopathologic features. Carcinogenesis. 2014 Oct; 35(10):2224-2231. doi: 10.1093/carcin/bgu127. [download]
- Barclay SS, Tamura T, Ito H, Fujita K, Tagawa K, Shimamura T, Katsuta A, Shiwaku H, Sone M, Imoto S, Miyano S, Okazawa H. Systems biology analysis of Drosophila in vivo screen data elucidates core networks for DNA damage repair in SCA1. Hum Mol Genet. 2014 Mar 1; 23(5):1345-1364. doi: 10.1093/hmg/ddt524. [download]
- Li C, Nagasaki M, Ikeda E, Sekiya Y, Miyano S. CSML2SBML: A novel tool for converting quantitative biological pathway models from CSML into SBML. Biosystems. 2014 Jul; 121:22-28. doi: 10.1016/j.biosystems.2014.05.004. [download]
- Damm F, Mylonas E, Cosson A, Yoshida K, Della Valle V, Mouly E, Diop M, Scourzic L, Shiraishi Y, Chiba K, Tanaka H, Miyano S, Kikushige Y, Davi F, Lambert J, Gautheret D, Merle-Béral H, Sutton L, Dessen P, Solary E, Akashi K, Vainchenker W, Mercher T, Droin N, Ogawa S, Nguyen-Khac F, Bernard OA. Acquired initiating mutations in early hematopoietic cells of CLL patients. Cancer Discov. 2014 Sep; 4(9):1088-1101. doi: 10.1158/2159-8290.CD-14-0104. [download]
- Haferlach T, Nagata Y, Grossmann V, Okuno Y, Bacher U, Nagae G, Schnittger S, Sanada M, Kon A, Alpermann T, Yoshida K, Roller A, Nadarajah N, Shiraishi Y, Shiozawa Y, Chiba K, Tanaka H, Koeffler HP, Klein HU, Dugas M, Aburatani H, Kohlmann A, Miyano S, Haferlach C, Kern W, Ogawa S. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia. 2014 Feb; 28(2):241-247. doi: 10.1038/leud.2013.336. [download]
- Hasegawa S, Imai K, Yoshida K, Okuno Y, Muramatsu H, Shiraishi Y, Chiba K, Tanaka H, Miyano S, Kojima S, Ogawa S, Morio T, Mizutani S, Takagi M. Whole-exome sequence analysis of ataxia telangiectasia-like phenotype. J Neurol Sci. 2014 May 15; 340(1-2):86-90. doi: 10.1016/j.jns.2014.02.033. [download]
- Hasegawa T, Mori T, Yamaguchi R, Imoto S, Miyano S, Akutsu T. An efficient data assimilation schema for restoration and extension of gene regulatory networks using time-course observation data. J Comput Biol. 2014 Nov; 21(11):785-798. doi: 10.1089/cmb.2014.0171. [download]
- Hasegawa T, Nagasaki M, Yamaguchi R, Imoto S, Miyano S. An efficient method of exploring simulation models by assimilating literature and biological observational data. Biosystems. 2014 Jul; 121:54-66. doi: 10.1016/j.Biosystems.2014.06.001. [download]
- Hasegawa T, Yamaguchi R, Nagasaki M, Miyano S, Imoto S. Inference of gene regulatory networks incorporating multi-source biological knowledge via a state space model with L1 regularization. PLoS One. 2014 Aug 27; 9(8):e105942. doi: 10.1371/journal.pone.0105942. [download]
- Hosono N, Makishima H, Jerez A, Yoshida K, Przychodzen B, McMahon S, Shiraishi Y, Chiba K, Tanaka H, Miyano S, Sanada M, Gómez-Seguí I, Verma AK, McDevitt MA, Sekeres MA, Ogawa S, Maciejewski JP. Recurrent genetic defects on chromosome 7q in myeloid neoplasms. Leukemia. 2014 Jun;28(6):1348-1351. doi: 10.1038/leu.2014.25. [download]
- Katayama K, Yamaguchi R, Imoto S, Watanabe K, Miyano S. Analysis of questionnaire for traditional medicine and development of decision support system. Evid Based Complement Alternat Med. 2014 Jan 29; 2014:974139. doi: 10.1155/2014/974139. [download]
- Kihara R, Nagata Y, Kiyoi H, Kato T, Yamamoto E, Suzuki K, Chen F, Asou N, Ohtake S, Miyawaki S, Miyazaki Y, Sakura T, Ozawa Y, Usui N, Kanamori H, Kiguchi T, Imai K, Uike N, Kimura F, Kitamura K, Nakaseko C, Onizuka M, Takeshita A, Ishida F, Suzushima H, Kato Y, Miwa H, Shiraishi Y, Chiba K, Tanaka H, Miyano S, Ogawa S, Naoe T. Comprehensive analysis of genetic alterations and their prognostic impacts in adult acute myeloid leukemia patients. Leukemia. 2014 Aug; 28(8):1586-1595. doi: 10.1038/leu.2014.55. [download]
- Matsunawa M, Yamamoto R, Sanada M, Sato-Otsubo A, Shiozawa Y, Yoshida K, Otsu M, Shiraishi Y, Miyano S, Isono K, Koseki H, Nakauchi H, Ogawa S. Haploinsufficiency of Sf3b1 leads to compromised stem cell function but not to myelodysplasia. Leukemia. 2014 Sep; 28(9):1844-1850. doi: 10.1038/leu.2014.73. [download]
- Oki T, Nishimura K, Kitaura J, Togami K, Maehara A, Izawa K, Sakaue-Sawano A, Niida A, Miyano S, Aburatani H, Kiyonari H, Miyawaki A, Kitamura T. A novel cell-cycle-indicator, mVenus-p27K-, identifies quiescent cells and visualizes G0-G1 transition. Sci Rep. 2014 Feb 6; 4:4012. doi: 10.1038/srep04012. [download]
- Park H, Shimamura T, Miyano S, Imoto S. Robust prediction of anti-cancer drug sensitivity and sensitivity-specific biomarker. PLoS One. 2014 Oct 17; 9(10):e108990. doi: 10.1371/journal.pone.0108990. [download]
- Saini H, Raicar G, Lal S, Dehzangi A, Lyons J, Paliwal KK, Imoto S, Miyano S, Sharma A. Genetic algorithm for an optimized weighted voting scheme incorporating k-separated bigram transition probabilities to improve protein fold recognition. 2014 Asia-Pacific World Congress on Computer Science and Engineering (APWC on CSE). 2014 Nov 4-5; 1: 1-7. DOI: 10.1109/APWCCSE.2014.7053846. https://www.computer.org/csdl/proceedings-article/apwc-on-cse/2014/07053846/12OmNAoDi8a [download]
- Saito MM, Imoto S, Yamaguchi R, Miyano S, Higuchi T. Parameter estimation in multi-compartment SIR model. 17th International Conference on Information Fusion (FUSION). 2014 Jul; 1-5. https://ieeexplore.ieee.org/document/6916269. [download]
- Sakata-Yanagimoto M, Enami T, Yoshida K, Shiraishi Y, Ishii R, Miyake Y, Muto H, Tsuyama N, Sato-Otsubo A, Okuno Y, Sakata S, Kamada Y, Nakamoto-Matsubara R, Tran NB, Izutsu K, Sato Y, Ohta Y, Furuta J, Shimizu S, Komeno T, Sato Y, Ito T, Noguchi M, Noguchi E, Sanada M, Chiba K, Tanaka H, Suzukawa K, Nanmoku T, Hasegawa Y, Nureki O, Miyano S, Nakamura N, Takeuchi K, Ogawa S, Chiba S. Somatic RHOA mutation in angioimmunoblastic T cell lymphoma. Nat Genet. 2014 Feb;46(2):171-175. doi: 10.1038/ng.2872. [download]
- Sato Y, Maekawa S, Ishii R, Sanada M, Morikawa T, Shiraishi Y, Yoshida K, Nagata Y, Sato-Otsubo A, Yoshizato T, Suzuki H, Shiozawa Y, Kataoka K, Kon A, Aoki K, Chiba K, Tanaka H, Kume H, Miyano S, Fukayama M, Nureki O, Homma Y, Ogawa S. Recurrent somatic mutations underlie corticotropin-independent Cushing's syndrome. Science. 2014 May 23; 344(6186):917-920. doi: 10.1126/science.1252328. [download]
- Sawada G, Takahashi Y, Niida A, Shimamura T, Kurashige J, Matsumura T, Ueo H, Uchi R, Takano Y, Ueda M, Hirata H, Sakimura S, Shinden Y, Eguchi H, Sudo T, Sugimachi K, Miyano S, Doki Y, Mori M, Mimori K. Loss of CDCP1 expression promotes invasiveness and poor prognosis in esophageal squamous cell carcinoma. Ann Surg Oncol. 2014 Dec; 21 Suppl 4:S640-7. doi: 10.1245/s10434-014-3740-4. [download]
- Seki M, Yoshida K, Shiraishi Y, Shimamura T, Sato Y, Nishimura R, Okuno Y, Chiba K, Tanaka H, Kato K, Kato M, Hanada R, Nomura Y, Park MJ, Ishida T, Oka A, Igarashi T, Miyano S, Hayashi Y, Ogawa S, Takita J. Biallelic DICER1 mutations in sporadic pleuropulmonary blastoma. Cancer Res. 2014 May 15; 74(10):2742-2749. doi: 10.1158/0008-5472.CAN-13-2470. [download]
- Sharma A, Dehzangi A, Lyons J, Imoto S, Miyano S, Nakai K, Patil A. Evaluation of sequence features from intrinsically disordered regions for the estimation of protein function. PLoS One. 2014 Feb 24; 9(2):e89890. doi: 10.1371/journal.pone.0089890. [download]
- Sharma A, Paliwal KK, Imoto S, Miyano S. A feature selection method using improved regularized linear discriminant analysis. Mach Vis Appl. 2014 Apr; 25(3): 775-786. doi: 10.1007/s00138-013-0577-y. [download]
- Sharma A, Paliwal KK, Imoto S, Miyano S, Sharma V, Ananthanarayanan R. A feature selection method using fixed-point algorithm for DNA microarray gene expression data. Int J Knowl Based Intell Eng Syst. 2014 Feb 27; 18(1): 55-59. doi: 10.3233/KES-140285. [download]
- Shen W, Clemente MJ, Hosono N, Yoshida K, Przychodzen B, Yoshizato T, Shiraishi Y, Miyano S, Ogawa S, Maciejewski JP, Makishima H. Deep sequencing reveals stepwise mutation acquisition in paroxysmal nocturnal hemoglobinuria. J Clin Invest. 2014 Oct; 124(10):4529-38. doi: 10.1172/JCI74747. [download]
- Sugimachi K, Niida A, Yamamoto K, Shimamura T, Imoto S, Iinuma H, Shinden Y, Eguchi H, Sudo T, Watanabe M, Tanaka J, Kudo S, Hase K, Kusunoki M, Yamada K, Shimada Y, Sugihara K, Maehara Y, Miyano S, Mori M, Mimori K. Allelic imbalance at an 8q24 oncogenic SNP is involved in activating MYC in human colorectal cancer. Ann Surg Oncol. 2014 Dec; 21 Suppl 4:S515-21. doi: 10.1245/s10434-013-3468-6. [download]
- Tokunaga H, Munakata K, Katayama K, Yamaguchi R, Imoto S, Miyano S, Watanabe K. Clinical data mining related to the Japanese kampo concept "hie" (oversensitivity to coldness) in men and pre- and postmenopausal women. Evid Based Complement Alternat Med. 2014 Feb 24; 2014:832824. doi: 10.1155/2014/832824. [download]
- Totoki Y, Yoshida A, Hosoda F, Nakamura H, Hama N, Ogura K, Yoshida A, Fujiwara T, Arai Y, Toguchida J, Tsuda H, Miyano S, Kawai A, Shibata T. Unique mutation portraits and frequent COL2A1 gene alteration in chondrosarcoma. Genome Res. 2014 Sep; 24(9):1411-1420. doi: 10.1101/gr.160598.113. [download]
- Usuyama N, Shiraishi Y, Sato Y, Kume H, Homma Y, Ogawa S, Miyano S, Imoto S. HapMuC: somatic mutation calling using heterozygous germ line variants near candidate mutations. Bioinformatics. 2014 Dec 1; 30(23):3302-3309. doi: 10.1093/Bioinformatics/btu537. [download]
- Yamaguchi K, Yamaguchi R, Takahashi N, Ikenoue T, Fujii T, Shinozaki M, Tsurita G, Hata K, Niida A, Imoto S, Miyano S, Nakamura Y, Furukawa Y. Overexpression of cohesion establishment factor DSCC1 through E2F in colorectal cancer. PLoS One. 2014 Jan 17; 9(1):e85750. doi: 10.1371/journal.pone.0085750. [download]
- Affara M, Sanders D, Araki H, Tamada Y, Dunmore BJ, Humphreys S, Imoto S, Savoie C, Miyano S, Kuhara S, Jeffries D, Print C, Charnock-Jones DS. Vasohibin-1 is identified as a master-regulator of endothelial cell apoptosis using gene network analysis. BMC Genomics. 2013 Jan 16; 14:23. doi: 10.1186/1471-2164-14-23. [download]
- Damm F, Chesnais V, Nagata Y, Yoshida K, Scourzic L, Okuno Y, Itzykson R, Sanada M, Shiraishi Y, Gelsi-Boyer V, Renneville A, Miyano S, Mori H, Shih LY, Park S, Dreyfus F, Guerci-Bresler A, Solary E, Rose C, Cheze S, Prébet T, Vey N, Legentil M, Duffourd Y, de Botton S, Preudhomme C, Birnbaum D, Bernard OA, Ogawa S, Fontenay M, Kosmider O. BCOR and BCORL1 mutations in myelodysplastic syndromes and related disorders. Blood. 2013 Oct 31; 122(18):3169-3177. doi: 10.1182/Blood-2012-11-469619. [download]
- Furuta M, Kozaki K, Tanimoto K, Tanaka S, Arii S, Shimamura T, Niida A, Miyano S, Inazawa J. The tumor-suppressive miR-497-195 cluster targets multiple cell-cycle regulators in hepatocellular carcinoma. PLoS One. 2013 Mar 27; 8(3):e60155. doi: 10.1371/journal.pone.0060155. [download]
- Gómez-Seguí I, Makishima H, Jerez A, Yoshida K, Przychodzen B, Miyano S, Shiraishi Y, Husseinzadeh HD, Guinta K, Clemente M, Hosono N, McDevitt MA, Moliterno AR, Sekeres MA, Ogawa S, Maciejewski JP. Novel recurrent mutations in the RAS-like GTP-binding gene RIT1 in myeloid malignancies. Leukemia. 2013 Sep;27(9):1943-1946. doi: 10.1038/leu.2013.179. [download]
- Hira A, Yabe H, Yoshida K, Okuno Y, Shiraishi Y, Chiba K, Tanaka H, Miyano S, Nakamura J, Kojima S, Ogawa S, Matsuo K, Takata M, Yabe M. Variant ALDH2 is associated with accelerated progression of bone marrow failure in Japanese Fanconi anemia patients. Blood. 2013 Oct 31;122(18):3206-3209. doi: 10.1182/Blood-2013-06-507962. [download]
- Ishikawa T, Shimizu T, Ueki A, Yamaguchi SI, Onishi N, Sugihara E, Kuninaka S, Miyamoto T, Morioka H, Nakayama R, Kobayashi E, Toyama Y, Mabuchi Y, Matsuzaki Y, Yamaguchi R, Miyano S, Saya H. Twist2 functions as a tumor suppressor in murine osteosarcoma cells. Cancer Sci. 2013 Jul; 104(7):880-888. doi: 10.1111/cas.12163. [download]
- Jeong E, Nagasaki M, Miyano S. Cell System Ontology. Encyclopedia of Systems Biology. Dubitzky, W., Wolkenhauer, O., Cho, KH., Yokota, H. (eds). 2013 Jun 05; 370-372. https://doi.org/10.1007/978-1-4419-9863-7_1109 https://link.springer.com/content/pdf/10.1007/978-1-4419-9863-7_1109.pdf Review [download]
- Katayama K, Yoshino T, Munakata K, Yamaguchi R, Imoto S, Miyano S, Watanabe K. Prescription of kampo drugs in the Japanese health care insurance program. Evid Based Complement Alternat Med. 2013 Nov 30; 2013:576973. doi: 10.1155/2013/576973. [download]
- Kayano M, Imoto S, Yamaguchi R, Miyano S. Multi-omics approach for estimating metabolic networks using low-order partial correlations. J Comput Biol. 2013 Aug; 20(8):571-582. doi: 10.1089/cmb.2013.0043. [download]
- Kitamura K, Yoshida K, Shiraishi Y, Chiba K, Tanaka H, Furukawa K, Miyano S, Ogawa S, Kunishima S. Normal neutrophil myosin IIA localization in an immunofluorescence analysis can rule out MYH9 disorders. J Thromb Haemost. 2013 Nov; 11(11):2071-2073. doi: 10.1111/jth.12406. [download]
- Komatsu M, Yoshimaru T, Matsuo T, Kiyotani K, Miyoshi Y, Tanahashi T, Rokutan K, Yamaguchi R, Saito A, Imoto S, Miyano S, Nakamura Y, Sasa M, Shimada M, Katagiri T. Molecular features of triple negative breast Cancer Cells by genome-wide gene expression profiling analysis. Int J Oncol. 2013 Feb; 42(2):478-506. doi: 10.3892/ijo.2012.1744. [download]
- Kon A, Shih LY, Minamino M, Sanada M, Shiraishi Y, Nagata Y, Yoshida K, Okuno Y, Bando M, Nakato R, Ishikawa S, Sato-Otsubo A, Nagae G, Nishimoto A, Haferlach C, Nowak D, Sato Y, Alpermann T, Nagasaki M, Shimamura T, Tanaka H, Chiba K, Yamamoto R, Yamaguchi T, Otsu M, Obara N, Sakata-Yanagimoto M, Nakamaki T, Ishiyama K, Nolte F, Hofmann WK, Miyawaki S, Chiba S, Mori H, Nakauchi H, Koeffler HP, Aburatani H, Haferlach T, Shirahige K, Miyano S, Ogawa S. Recurrent mutations in multiple components of the cohesin complex in myeloid neoplasms. Nat Genet. 2013 Oct; 45(10):1232-1237. doi: 10.1038/ng.2731. [download]
- Kunishima S, Okuno Y, Yoshida K, Shiraishi Y, Sanada M, Muramatsu H, Chiba K, Tanaka H, Miyazaki K, Sakai M, Ohtake M, Kobayashi R, Iguchi A, Niimi G, Otsu M, Takahashi Y, Miyano S, Saito H, Kojima S, Ogawa S. ACTN1 mutations cause congenital macrothrombocytopenia. Am J Hum Genet. 2013 Mar 7; 92(3):431-438. doi: 10.1016/j.ajhg.2013.01.015. [download]
- Lin TL, Nagata Y, Kao HW, Sanada M, Okuno Y, Huang CF, Liang DC, Kuo MC, Lai CL, Lee EH, Shih YS, Tanaka H, Shiraishi Y, Chiba K, Lin TH, Wu JH, Miyano S, Ogawa S, Shih LY. Clonal leukemic evolution in myelodysplastic syndromes with TET2 and IDH1/2 mutations. Haematologica. 2014 Jan; 99(1):28-36. doi: 10.3324/haematol.2013.091249. [download]
- Makishima H, Yoshida K, Nguyen N, Przychodzen B, Sanada M, Okuno Y, Ng KP, Gudmundsson KO, Vishwakarma BA, Jerez A, Gomez-Segui I, Takahashi M, Shiraishi Y, Nagata Y, Guinta K, Mori H, Sekeres MA, Chiba K, Tanaka H, Muramatsu H, Sakaguchi H, Paquette RL, McDevitt MA, Kojima S, Saunthararajah Y, Miyano S, Shih LY, Du Y, Ogawa S, Maciejewski JP. Somatic SETBP1 mutations in myeloid malignancies. Nat Genet. 2013 Aug; 45(8):942-946. doi: 10.1038/ng.2696. [download]
- Nagasaki M, Fujita A, Sekiya Y, Saito A, Ikeda E, Li C, Miyano S. XiP: a computational environment to create, extend and share workflows. Bioinformatics. 2013 Jan 1; 29(1):137-139. doi: 10.1093/Bioinformatics/bts630. [download]
- Niida A, Tremmel G, Imoto S, Miyano S. Multilayer Cluster Heat Map Visualizing Biological Tensor Data. Lecture Notes in Bioinformatics. 2013 Nov; 8213: 116-125. doi: 10.1007/978-3-319-02624-4_11. [download]
- Osawa T, Tsuchida R, Muramatsu M, Shimamura T, Wang F, Suehiro J, Kanki Y, Wada Y, Yuasa Y, Aburatani H, Miyano S, Minami T, Kodama T, Shibuya M. Inhibition of histone demethylase JMJD1A improves anti-angiogenic therapy and reduces tumor-associated macrophages. Cancer Res. 2013 May 15; 73(10):3019-3028. doi: 10.1158/0008-5472.CAN-12-3231. [download]
- Saida S, Watanabe K, Sato-Otsubo A, Terui K, Yoshida K, Okuno Y, Toki T, Wang R, Shiraishi Y, Miyano S, Kato I, Morishima T, Fujino H, Umeda K, Hiramatsu H, Adachi S, Ito E, Ogawa S, Ito M, Nakahata T, Heike T. Clonal selection in xenografted TAM recapitulates the evolutionary process of myeloid leukemia in Down syndrome. Blood. 2013 May 23; 121(21):4377-4387. doi: 10.1182/Blood-2012-12-474387. [download]
- Saito MM, Imoto S, Yamaguchi R, Miyano S, Higuchi T. Estimation of abrupt changes in sentinel observation data of influenza epidemics in Japan. Proceedings of the 16th International Conference on Information Fusion. 2013 Jul; 1385-1390. https://ieeexplore.ieee.org/document/6641160. [download]
- Saito MM, Imoto S, Yamaguchi R, Sato H, Nakada H, Kami M, Miyano S, Higuchi T. Extension and verification of the SEIR model on the 2009 influenza A (H1N1) pandemic in Japan. Math Biosci. 2013 Nov; 246(1):47-54. doi: 10.1016/j.mbs.2013.08.009. [download]
- Saito MM, Imoto S, Yamaguchi R, Tsubokura M, Kami M, Nakada H, Sato H, Miyano S, Higuchi T. Enhancement of collective immunity in Tokyo metropolitan area by selective vaccination against an emerging influenza pandemic. PLoS One. 2013 Sep 18; 8(9):e72866. doi: 10.1371/journal.pone.0072866. [download]
- Sakaguchi H, Okuno Y, Muramatsu H, Yoshida K, Shiraishi Y, Takahashi M, Kon A, Sanada M, Chiba K, Tanaka H, Makishima H, Wang X, Xu Y, doi:saki S, Hama A, Nakanishi K, Takahashi Y, Yoshida N, Maciejewski JP, Miyano S, Ogawa S, Kojima S. Exome sequencing identifies secondary mutations of SETBP1 and JAK3 in juvenile myelomonocytic leukemia. Nat Genet. 2013 Aug; 45(8):937-941. doi: 10.1038/ng.2698. [download]
- Sato Y, Yoshizato T, Shiraishi Y, Maekawa S, Okuno Y, Kamura T, Shimamura T, Sato-Otsubo A, Nagae G, Suzuki H, Nagata Y, Yoshida K, Kon A, Suzuki Y, Chiba K, Tanaka H, Niida A, Fujimoto A, Tsunoda T, Morikawa T, Maeda D, Kume H, Sugano S, Fukayama M, Aburatani H, Sanada M, Miyano S, Homma Y, Ogawa S. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat Genet. 2013 Aug; 45(8):860-867. doi: 10.1038/ng.2699. [download]
- Sharma A, Paliwal KK, Dehzangi A, Lyons J, Imoto S, Miyano S. A strategy to select suitable physicochemical attributes of amino acids for protein fold recognition. BMC Bioinformatics. 2013 Jul 24; 14:233. doi: 10.1186/1471-2105-14-233. [download]
- Sharma A, Paliwal KK, Imoto S, Miyano S. Principal component analysis using QR decomposition. Int J Mach Learn Cybern. 2013 Dec; 4(6): 679-683. doi: 10.1007/s13042-012-0131-7. [download]
- Shiraishi Y, Sato Y, Chiba K, Okuno Y, Nagata Y, Yoshida K, Shiba N, Hayashi Y, Kume H, Homma Y, Sanada M, Ogawa S, Miyano S. An empirical Bayesian framework for somatic mutation detection from cancer genome sequencing data. Nucleic Acids Res. 2013 Apr; 41(7):e89. doi: 10.1093/nar/gkt126. [download]
- Takatsuno Y, Mimori K, Yamamoto K, Sato T, Niida A, Inoue H, Imoto S, Kawano S, Yamaguchi R, Toh H, Iinuma H, Ishimaru S, Ishii H, Suzuki S, Tokudome S, Watanabe M, Tanaka J, Kudo SE, Mochizuki H, Kusunoki M, Yamada K, Shimada Y, Moriya Y, Miyano S, Sugihara K, Mori M. The rs6983267 SNP is associated with MYC transcription efficiency, which promotes progression and worsens prognosis of colorectal cancer. Ann Surg Oncol. 2013 Apr; 20(4):1395-1402. doi: 10.1245/s10434-012-2657-z. [download]
- Tamura T, Sone M, Nakamura Y, Shimamura T, Imoto S, Miyano S, Okazawa H. A restricted level of PQBP1 is needed for the best longevity of Drosophila. Neurobiol Aging. 2013 Jan; 34(1):356.e11-20. doi: 10.1016/j.neurobiolaging.2012.07.015. [download]
- Yamaguchi R, Imoto S, Kami M, Watanabe K, Miyano S, Yuji K. Does Twitter trigger bursts in signature collections? PLoS One. 2013 Mar 6; 8(3):e58252. doi: 10.1371/journal.pone.0058252. [download]
- Yokobori T, Iinuma H, Shimamura T, Imoto S, Sugimachi K, Ishii H, Iwatsuki M, Ota D, Ohkuma M, Iwaya T, Nishida N, Kogo R, Sudo T, Tanaka F, Shibata K, Toh H, Sato T, Barnard GF, Fukagawa T, Yamamoto S, Nakanishi H, Sasaki S, Miyano S, Watanabe T, Kuwano H, Mimori K, Pantel K, Mori M. Plastin3 is a novel marker for circulating tumor cells undergoing the epithelial-mesenchymal transition and is associated with colorectal cancer prognosis. Cancer Res. 2013 Apr 1; 73(7):2059-2069. doi: 10.1158/0008-5472.CAN-12-0326. [download]
- Yoshida K, Toki T, Okuno Y, Kanezaki R, Shiraishi Y, Sato-Otsubo A, Sanada M, Park MJ, Terui K, Suzuki H, Kon A, Nagata Y, Sato Y, Wang R, Shiba N, Chiba K, Tanaka H, Hama A, Muramatsu H, Hasegawa D, Nakamura K, Kanegane H, Tsukamoto K, Adachi S, Kawakami K, Kato K, Nishimura R, Izraeli S, Hayashi Y, Miyano S, Kojima S, Ito E, Ogawa S. The landscape of somatic mutations in Down syndrome-related myeloid disorders. Nat Genet. 2013 Nov; 45(11):1293-1299. doi: 10.1038/ng.2759. Erratum in: Nat Genet. 2013 Dec; 45(12):1516. doi: 10.1038/ng1213-1516. [download]
- Yoshimaru T, Komatsu M, Matsuo T, Chen YA, Murakami Y, Mizuguchi K, Mizohata E, Inoue T, Akiyama M, Yamaguchi R, Imoto S, Miyano S, Miyoshi Y, Sasa M, Nakamura Y, Katagiri T. Targeting BIG3-PHB2 interaction to overcome tamoxifen resistance in breast Cancer Cells. Nat Commun. 2013 Sep 20; 4:2443. doi: 10.1038/ncomms3443. [download]
- Yoshino T, Katayama K, Munakata K, Horiba Y, Yamaguchi R, Imoto S, Miyano S, Watanabe K. Statistical analysis of hie (cold sensation) and hiesho (cold disorder) in kampo clinic. Evid Based Complement Alternat Med. 2013 Dec 30; 2013:398458. doi: 10.1155/2013/398458. [download]
- Fujimori S, Hino K, Saito A, Miyano S, Miyamoto-Sato E. PRD: A protein-RNA interaction database. Bioinformation. 2012 Aug 3; 8(15):729-730. doi: 10.6026/97320630008729. [download]
- Fujimori S, Hirai N, Masuoka K, Oshikubo T, Yamashita T, Washio T, Saito A, Nagasaki M, Miyano S, Miyamoto-Sato E. IRView: a database and viewer for protein interacting regions. Bioinformatics. 2012 Jul 15; 28(14):1949-50. doi: 10.1093/Bioinformatics/bts289. [download]
- Fujimoto A, Totoki Y, Abe T, Boroevich KA, Hosoda F, Nguyen HH, Aoki M, Hosono N, Kubo M, Miya F, Arai Y, Takahashi H, Shirakihara T, Nagasaki M, Shibuya T, Nakano K, Watanabe-Makino K, Tanaka H, Nakamura H, Kusuda J, Ojima H, Shimada K, Okusaka T, Ueno M, Shigekawa Y, Kawakami Y, Arihiro K, Ohdan H, Gotoh K, Ishikawa O, Ariizumi S, Yamamoto M, Yamada T, Chayama K, Kosuge T, Yamaue H, Kamatani N, Miyano S, Nakagama H, Nakamura Y, Tsunoda T, Shibata T, Nakagawa H. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet. 2012 May 27; 44(7):760-764. doi: 10.1038/ng.2291. [download]
- Fujita A, Severino P, Kojima K, Sato JR, Patriota AG, Miyano S. Functional clustering of time series gene expression data by Granger causality. BMC Syst Biol. 2012 Oct 30; 6:137. doi: 10.1186/1752-0509-6-137. [download]
- Hurley D, Araki H, Tamada Y, Dunmore B, Sanders D, Humphreys S, Affara M, Imoto S, Yasuda K, Tomiyasu Y, Tashiro K, Savoie C, Cho V, Smith S, Kuhara S, Miyano S, Charnock-Jones DS, Crampin EJ, Print CG. Gene network inference and visualization tools for biologists: application to new human transcriptome datasets. Nucleic Acids Res. 2012 Mar;40(6):2377-2398. doi: 10.1093/nar/gkr902. [download]
- Ishimaru S, Mimori K, Yamamoto K, Inoue H, Imoto S, Kawano S, Yamaguchi R, Sato T, Toh H, Iinuma H, Maeda T, Ishii H, Suzuki S, Tokudome S, Watanabe M, Tanaka J, Kudo SE, Sugihara K, Hase K, Mochizuki H, Kusunoki M, Yamada K, Shimada Y, Moriya Y, Barnard GF, Miyano S, Mori M. Increased risk for CRC in diabetic patients with the nonrisk allele of SNPs at 8q24. Ann Surg Oncol. 2012 Sep; 19(9):2853-2858. doi: 10.1245/s10434-012-2278-6. [download]
- Kawano S, Shimamura T, Niida A, Imoto S, Yamaguchi R, Nagasaki M, Yoshida R, Print C, Miyano S. Identifying gene pathways associated with cancer characteristics via sparse statistical methods. IEEE/ACM Trans Comput Biol Bioinform. 2012 Jul-Aug; 9(4):966-972. doi: 10.1109/TCBB.2012.48. https://dl.acm.org/doi/10.1109/TCBB.2012.48. [download]
- Kojima K, Imoto S, Yamaguchi R, Fujita A, Yamauchi M, Gotoh N, Miyano S. Identifying regulational alterations in gene regulatory networks by state space representation of vector autoregressive models and variational annealing. BMC Genomics. 2012 Jan 17;13 Suppl 1(Suppl 1):S6. doi: 10.1186/1471-2164-13-S1-S6. [download]
- Niida A, Imoto S, Shimamura T, Miyano S. Statistical model-based testing to evaluate the recurrence of genomic aberrations. Bioinformatics. 2012 Jun 15; 28(12):i115-120. doi: 10.1093/Bioinformatics/bts203. [download]
- Ogami K, Yamaguchi R, Imoto S, Tamada Y, Araki H, Print C, Miyano S. Computational gene network analysis reveals TNF-induced angiogenesis. BMC Syst Biol. 2012 Dec 12; 6 Suppl 2(Suppl 2):S12. doi: 10.1186/1752-0509-6-S2-S12. [download]
- Okayama H, Kohno T, Ishii Y, Shimada Y, Shiraishi K, Iwakawa R, Furuta K, Tsuta K, Shibata T, Yamamoto S, Watanabe S, Sakamoto H, Kumamoto K, Takenoshita S, Gotoh N, Mizuno H, Sarai A, Kawano S, Yamaguchi R, Miyano S, Yokota J. Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res. 2012 Jan 1;72(1):100-111. doi: 10.1158/0008-5472.CAN-11-1403. [download]
- Saito MM, Imoto S, Yamaguchi R, Miyano S, Higuchi T. Identifiability of local transmissibility parameters in agent-based pandemic simulation. 2012 15th International Conference on Information Fusion. 2012 Jul; 2466-2471. https://ieeexplore.ieee.org/document/6290603. [download]
- Sharma A, Imoto S, Miyano S. A between-class overlapping filter-based method for transcriptome data analysis. J Bioinform Comput Biol. 2012 Oct; 10(5):1250010. doi: 10.1142/S0219720012500102. [download]
- Sharma A, Imoto S, Miyano S. A top-r feature selection algorithm for microarray gene expression data. IEEE/ACM Trans Comput Biol Bioinform. 2012 May-Jun; 9(3):754-764. doi: 10.1109/TCBB.2011.151. [download]
- Sharma A, Imoto S, Miyano S, Sharma V. Null space based feature selection method for gene expression data. Int J Mach Learn Cybern. 2012 Dec; 3(4): 269-276. doi: 10.1007/s13042-011-0061-9. [download]
- Wang L, Hurley DG, Watkins W, Araki H, Tamada Y, Muthukaruppan A, Ranjard L, Derkac E, Imoto S, Miyano S, Crampin EJ, Print CG. Cell cycle gene networks are associated with melanoma prognosis. PLoS One. 2012 Apr 20; 7(4):e34247. doi: 10.1371/journal.pone.0034247. [download]
- Yamamoto M, Yamaguchi R, Munakata K, Takashima K, Nishiyama M, Hioki K, Ohnishi Y, Nagasaki M, Imoto S, Miyano S, Ishige A, Watanabe K. A microarray analysis of gnotobiotic mice indicating that microbial exposure during the neonatal period plays an essential role in immune system development. BMC Genomics. 2012 Jul 23; 13:335. doi: 10.1186/1471-2164-13-335. [download]
- Yamauchi M, Yamaguchi R, Nakata A, Kohno T, Nagasaki M, Shimamura T, Imoto S, Saito A, Ueno K, Hatanaka Y, Yoshida R, Higuchi T, Nomura M, Beer DG, Yokota J, Miyano S, Gotoh N. Epidermal growth factor receptor tyrosine kinase defines critical prognostic genes of stage I lung adenocarcinoma. PLoS One. 2012 Sep 9; 7(9):e43923. doi: 10.1371/journal.pone.0043923. [download]
- Yasuda T, Suzuki S, Nagasaki M, Miyano S. ChopSticks: High-resolution analysis of homozygous deletions by exploiting concordant read pairs. BMC Bioinformatics. 2012 Oct 30; 13:279. doi: 10.1186/1471-2105-13-279. [download]
- Katayama K, Imoto S, Watanabe K, Ymaguchi R, Matsuura K, Miyano S. Analysis of questionnaire for traditional medical and develop decision support system. 2012 IEEE International Conference on Bioinformatics and Biomedicine Workshops (BIBMW). 2012 Oct 4-7; 762-763. https://ieeexplore.ieee.org/document/6470235 [download]
- Chalkidis G, Nagasaki M, Miyano S. High performance hybrid functional Petri net simulations of biological pathway models on CUDA. IEEE/ACM Trans Comput Biol Bioinform. 2011 Nov-Dec;8(6):1545-1556. doi: 10.1109/TCBB.2010.118. https://dl.acm.org/doi/10.1109/TCBB.2010.118. [download]
- Fujita A, Sato JR, Angelo M, Demasi A, Yamaguchi R, Shimamura T, Ferreira CE, Sogayar MC, Miyano S. Inferring contagion in regulatory networks. IEEE/ACM Trans Comput Biol Bioinform. 2011 Mar-Apr; 8(2):570-576. doi: 10.1109/TCBB.2010.40. https://dl.acm.org/doi/10.1109/TCBB.2010.40. [download]
- Hasegawa T, Yamaguchi R, Nagasaki M, Imoto S, Miyano S. Comprehensive Pharmacogenomic Pathway Screening by Data Assimilation. Lecture Notes in Bioinformatics. 2011 May: 6674: 160-171. doi: 10.1007/978-3-642-21260-4_18. [download]
- Imoto S, Tamada Y, Araki H, Miyano S. Computational Drug Target Pathway Discovery: A Bayesian Network Approach. H Lu, B Schokop, H Zhao (eds.) Handbook of Computational Statistics: Statistical Bioinformatics, Springer. 2011 Jan 01; 501-532. https://link.springer.com/chapter/10.1007/978-3-642-16345-6_24#citeas [download]
- Jeong E, Nagasaki M, Ikeda E, Sekiya Y, Saito A, Miyano S. CSO validator: improving manual curation workflow for biological pathways. Bioinformatics. 2011 Sep 1; 27(17):2471-2472. doi: 10.1093/Bioinformatics/btr395. [download]
- Jeong E, Nagasaki M, Ueno K, Miyano S. Ontology-based instance data validation for high-quality curated biological pathways. BMC Bioinformatics. 2011 Feb 15; 12 Suppl 1(Suppl 1):S8. doi: 10.1186/1471-2105-12-S1-S8. [download]
- Katayama K, Yamaguchi R, Imoto S, Matsuura K, Watanabe K, Miyano S. Clustering for Visual Analogue Scale Data in Symbolic Data Analysis. Procedia Computer Science. 2011 Oct 11; 6: 370-374. doi: 10.1016/j.procs.2011.08.068. [download]
- Katayama K, Yamaguchi R, Imoto S, Matsuura K, Watanabe K, Miyano S. Transform of visual analogue scale data and their clustering. Int J Knowl Eng Soft Data Paradigms. 2001 Mar 2; 3(2): 143-151. doi: 10.1504/IJKESDP.2011.045726. [download]
- Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, Miyano S, Mori M. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011 Oct 15; 71(20):6320-6326. doi: 10.1158/0008-5472.CAN-11-1021. Erratum in: Cancer Res. 2012 Feb 15; 72(4):1039. doi: 10.1158/0008-5472.CAN-12-0034. [download]
- Koh CH, Nagasaki M, Saito A, Li C, Wong L, Miyano S. MIRACH: efficient model checker for quantitative biological pathway models. Bioinformatics. 2011 Mar 1; 27(5):734-735. doi: 10.1093/Bioinformatics/btq727. [download]
- Li C, Nagasaki M, Koh CH, Miyano S. Online model checking approach based parameter estimation to a neuronal fate decision simulation model in Caenorhabditis elegans with hybrid functional Petri net with extension. Mol Biosyst. 2011 May; 7(5):1576-92. doi: 10.1039/c0mb00253d. [download]
- Matsuno H, Nagasaki M, Miyano S. Hybrid Petri net based modeling for biological pathway simulation. Natural Comput. 2011 Sep; 10(3): 1099-1120. doi: 10.1007/s11047-009-9164-6. [download]
- Mitou N, Matsuno H, Miyano S, Inouye ST. A Case Study of HFPN Simulation: Finding Essential Roles of Ror Gene in the Interaction of Feedback Loops in Mammalian Circadian Clock. Modeling in Systems Biology. 2011 Jan 1; Computational Biology 16: 281-306. doi: 10.1007/978-1-84996-474-6_13. [download]
- Nagasaki M, Saito A, Fujita A, Tremmel G, Ueno K, Ikeda E, Jeong E, Miyano S. Systems biology model repository for macrophage pathway simulation. Bioinformatics. 2011 Jun 1;27(11):1591-1593. doi: 10.1093/Bioinformatics/btr173. [download]
- Osoda T, Miyano S. 2D-Qsar for 450 types of amino acid induction peptides with a novel substructure pair descriptor having wider scope. J Cheminform. 2011 Nov 2; 3(1):50. doi: http://dx.doi.org/10.1186/1758-2946-3-50[download]
- Saito A, Nagasaki M, Matsuno H, Miyano S. Hybrid Functional Petri Net with Extension for Dynamic Pathway Modeling. Modeling in Systems Biology. 2011 Jan 1; Computational Biology 16: 101-120. doi: 10.1007/978-1-84996-474-6_6. [download]
- Saito MM, Imoto S, Yamaguchi R, Miyano S, Higuchi T. Estimation of macroscopic parameter in agent-based pandemic simulation. 14th International Conference on Information Fusion. 2011 Jul; 1-6. https://ieeexplore.ieee.org/document/5977691. [download]
- Saito MM, Imoto S, Yamaguchi R, Miyano S, Higuchi T. Parallel Agent-Based Simulator for Influenza Pandemic. Lecture Notes in Artificial Intelligence. 2011 May; 7068: 361-370. doi: 10.1007/978-3-642-27216-5_27. [download]
- Shimamura T, Imoto S, Shimada Y, Hosono Y, Niida A, Nagasaki M, Yamaguchi R, Takahashi T, Miyano S. A novel network profiling analysis reveals system changes in epithelial-mesenchymal transition. PLoS One. 2011 Jun 7; 6(6):e20804. doi: 10.1371/journal.pone.0020804. [download]
- Shiraishi Y, Okada-Hatakeyama M, Miyano S. A rank-based statistical test for measuring synergistic effects between two gene sets. Bioinformatics. 2011 Sep 1; 27(17):2399-2405. doi: 10.1093/Bioinformatics/btr382. [download]
- Suzuki S, Yasuda T, Shiraishi Y, Miyano S, Nagasaki M. ClipCrop: a tool for detecting structural variations with single-base resolution using soft-clipping information. BMC Bioinformatics. 2011 Dec 14; 12 Suppl 14(Suppl 14):S7. doi: 10.1186/1471-2105-12-S14-S7. [download]
- Tamada Y, Imoto S, Araki H, Nagasaki M, Print C, Charnock-Jones DS, Miyano S. Estimating genome-wide gene networks using nonparametric Bayesian network models on massively parallel computers. IEEE/ACM Trans Comput Biol Bioinform. 2011 May-Jun; 8(3):683-97. doi: 10.1109/TCBB.2010.68. https://dl.acm.org/doi/10.1109/TCBB.2010.68. [download]
- Tamada Y, Imoto S, Miyano S. Parallel Algorithm for Learning Optimal Bayesian Network Structure. J Mach Learn Res. 2011 Jul; 12: 2437-2459. https://dl.acm.org/doi/10.5555/1953048.2021080. [download]
- Tamada Y, Shimamura T, Yamaguchi R, Imoto S, Nagasaki M, Miyano S. SIGN: large-scale gene network estimation environment for high performance computing. Genome Informatics. 2011 Dec; 25(1):40-52. doi: 10.11234/gi.25.40. [download]
- Tamada Y, Yamaguchi R, Imoto S, Hirose O, Yoshida R, Nagasaki M, Miyano S. SiGN-SSM: Open source parallel software for estimating gene networks with state space models. Bioinformatics. 2011 Apr 15; 27(8):1172-1173. doi: 10.1093/Bioinformatics/btr078. [download]
- Yamaguchi K, Sakai M, Kim J, Tsunesumi S, Fujii T, Ikenoue T, Yamada Y, Akiyama Y, Muto Y, Yamaguchi R, Miyano S, Nakamura Y, Furukawa Y. MRG-binding protein contributes to colorectal cancer development. Cancer Sci. 2011 Aug; 102(8):1486-1492. doi: 10.1111/j.1349-7006.2011.01971.x. [download]
- Yamauchi M, Yoshino I, Yamaguchi R, Shimamura T, Nagasaki M, Imoto S, Niida A, Koizumi F, Kohno T, Yokota J, Miyano S, Gotoh N. N-cadherin expression is a potential survival mechanism of gefitinib-resistant lung Cancer Cells. Am J Cancer Res. 2011 Aug 8;1(7):823-833. PMC3196281 [download]
- Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, Sato Y, Sato-Otsubo A, Kon A, Nagasaki M, Chalkidis G, Suzuki Y, Shiosaka M, Kawahata R, Yamaguchi T, Otsu M, Obara N, Sakata-Yanagimoto M, Ishiyama K, Mori H, Nolte F, Hofmann WK, Miyawaki S, Sugano S, Haferlach C, Koeffler HP, Shih LY, Haferlach T, Chiba S, Nakauchi H, Miyano S, Ogawa S. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011 Sep 11; 478(7367):64-69. doi: 10.1038/nature10496. [download]
- Do JH, Nagasaki M, Miyano S. The systems approach to the prespore-specific activation of sigma factor SigF in Bacillus subtilis. Biosystems. 2010 Jun; 100(3):178-184. doi: 10.1016/j.Biosystems.2010.03.002. [download]
- Fujimoto A, Nakagawa H, Hosono N, Nakano K, Abe T, Boroevich KA, Nagasaki M, Yamaguchi R, Shibuya T, Kubo M, Miyano S, Nakamura Y, Tsunoda T. Whole-genome sequencing and comprehensive variant analysis of a Japanese individual using massively parallel sequencing. Nat Genet. 2010 Nov; 42(11):931-936. doi: 10.1038/ng.691. [download]
- Fujita A, Kojima K, Patriota AG, Sato JR, Severino P, Miyano S. A fast and robust statistical test based on likelihood ratio with Bartlett correction to identify Granger causality between gene sets. Bioinformatics. 2010 Sep 15;26(18):2349-2351. doi: 10.1093/Bioinformatics/btq427. [download]
- Fujita A, Nagasaki M, Imoto S, Saito A, Ikeda E, Shimamura T, Yamaguchi R, Hayashizaki Y, Miyano S. Comparison of gene expression profiles produced by CAGE, Illumina microarray and real time RT-PCR. Genome Informatics. 2010 Jul; 24:56-68. DOI: 10.1142/9781848166585_0005. [download]
- Fujita A, Sato JR, Demasi MAA, Miyano S, Sogayar MC, Ferreira CE. An introduction to time-varying connectivity estimation for gene regulatory networks. Frank Emmert-Streib, Matthias Dehmer (eds.) Medical Biostatistics for Complex Diseases. Weinheim, Germany. Wiley VCH Verlag. 2010 Apr 22; 205-230. https://onlinelibrary.wiley.com/doi/epdf/10.1002/9783527630332.ch11#accessDenialLayout Review [download]
- Fujita A, Severino P, Ricardo Sato J, Miyano S. Granger Causality in Systems Biology: Modeling Gene Networks in Time Series Microarray Data Using Vector Autoregressive Models. Lecture Notes in Bioinformatics. 2010 Aug-Sep; 6268: 13-24. doi: 10.1007/978-3-642-15060-9_2. [download]
- Higashigaki T, Kojima K, Yamaguchi R, Inoue M, Imoto S, Miyano S. Identifying Hidden Confounders in Gene Networks by Bayesian Networks. 2010 IEEE International Conference on BioInformatics and BioEngineering. 2010 May; 1: 168-173. doi: 10.1109/BIBE.2010.35. [download]
- Imoto S, Kojima K, Perrier E, Tamada Y, Miyano S. Searching Optimal Bayesian Network Structure on Constraint Search Space: Super-Structure Approach. Lecture Notes in Artificial Intelligence. 2010 Nov; 6797: 210-218. doi: 10.1007/978-3-642-25655-4_19. [download]
- International Cancer Genome Consortium, Hudson TJ, Anderson W, Artez A, Barker AD, Bell C, Bernabé RR, Bhan MK, Calvo F, Eerola I, Gerhard DS, Guttmacher A, Guyer M, Hemsley FM, Jennings JL, Kerr D, Klatt P, Kolar P, Kusada J, Lane DP, Laplace F, Youyong L, Nettekoven G, Ozenberger B, Peterson J, Rao TS, Remacle J, Schafer AJ, Shibata T, Stratton MR, Vockley JG, Watanabe K, Yang H, Yuen MM, Knoppers BM, Bobrow M, Cambon-Thomsen A, Dressler LG, Dyke SO, Joly Y, Kato K, Kennedy KL, Nicolás P, Parker MJ, Rial-Sebbag E, Romeo-Casabona CM, Shaw KM, Wallace S, Wiesner GL, Zeps N, Lichter P, Biankin AV, Chabannon C, Chin L, Clément B, de Alava E, Degos F, Ferguson ML, Geary P, Hayes DN, Hudson TJ, Johns AL, Kasprzyk A, Nakagawa H, Penny R, Piris MA, Sarin R, Scarpa A, Shibata T, van de Vijver M, Futreal PA, Aburatani H, Bayés M, Botwell DD, Campbell PJ, Estivill X, Gerhard DS, Grimmond SM, Gut I, Hirst M, López-Otín C, Majumder P, Marra M, McPherson JD, Nakagawa H, Ning Z, Puente XS, Ruan Y, Shibata T, Stratton MR, Stunnenberg HG, Swerdlow H, Velculescu VE, Wilson RK, Xue HH, Yang L, Spellman PT, Bader GD, Boutros PC, Campbell PJ, Flicek P, Getz G, Guigó R, Guo G, Haussler D, Heath S, Hubbard TJ, Jiang T, Jones SM, Li Q, López-Bigas N, Luo R, Muthuswamy L, Ouellette BF, Pearson JV, Puente XS, Quesada V, Raphael BJ, Sander C, Shibata T, Speed TP, Stein LD, Stuart JM, Teague JW, Totoki Y, Tsunoda T, Valencia A, Wheeler DA, Wu H, Zhao S, Zhou G, Stein LD, Guigó R, Hubbard TJ, Joly Y, Jones SM, Kasprzyk A, Lathrop M, López-Bigas N, Ouellette BF, Spellman PT, Teague JW, Thomas G, Valencia A, Yoshida T, Kennedy KL, Axton M, Dyke SO, Futreal PA, Gerhard DS, Gunter C, Guyer M, Hudson TJ, McPherson JD, Miller LJ, Ozenberger B, Shaw KM, Kasprzyk A, Stein LD, Zhang J, Haider SA, Wang J, Yung CK, Cros A, Liang Y, Gnaneshan S, Guberman J, Hsu J, Bobrow M, Chalmers DR, Hasel KW, Joly Y, Kaan TS, Kennedy KL, Knoppers BM, Lowrance WW, Masui T, Nicolás P, Rial-Sebbag E, Rodriguez LL, Vergely C, Yoshida T, Grimmond SM, Biankin AV, Bowtell DD, Cloonan N, deFazio A, Eshleman JR, Etemadmoghadam D, Gardiner BB, Kench JG, Scarpa A, Sutherland RL, Tempero MA, Waddell NJ, Wilson PJ, McPherson JD, Gallinger S, Tsao MS, Shaw PA, Petersen GM, Mukhopadhyay D, Chin L, DePinho RA, Thayer S, Muthuswamy L, Shazand K, Beck T, Sam M, Timms L, Ballin V, Lu Y, Ji J, Zhang X, Chen F, Hu X, Zhou G, Yang Q, Tian G, Zhang L, Xing X, Li X, Zhu Z, Yu Y, Yu J, Yang H, Lathrop M, Tost J, Brennan P, Holcatova I, Zaridze D, Brazma A, Egevard L, Prokhortchouk E, Banks RE, Uhlén M, Cambon-Thomsen A, Viksna J, Ponten F, Skryabin K, Stratton MR, Futreal PA, Birney E, Borg A, Børresen-Dale AL, Caldas C, Foekens JA, Martin S, Reis-Filho JS, Richardson AL, Sotiriou C, Stunnenberg HG, Thoms G, van de Vijver M, van't Veer L, Calvo F, Birnbaum D, Blanche H, Boucher P, Boyault S, Chabannon C, Gut I, Masson-Jacquemier JD, Lathrop M, Pauporté I, Pivot X, Vincent-Salomon A, Tabone E, Theillet C, Thomas G, Tost J, Treilleux I, Calvo F, Bioulac-Sage P, Clément B, Decaens T, Degos F, Franco D, Gut I, Gut M, Heath S, Lathrop M, Samuel D, Thomas G, Zucman-Rossi J, Lichter P, Eils R, Brors B, Korbel JO, Korshunov A, Landgraf P, Lehrach H, Pfister S, Radlwimmer B, Reifenberger G, Taylor MD, von Kalle C, Majumder PP, Sarin R, Rao TS, Bhan MK, Scarpa A, Pederzoli P, Lawlor RA, Delledonne M, Bardelli A, Biankin AV, Grimmond SM, Gress T, Klimstra D, Zamboni G, Shibata T, Nakamura Y, Nakagawa H, Kusada J, Tsunoda T, Miyano S, Aburatani H, Kato K, Fujimoto A, Yoshida T, Campo E, López-Otín C, Estivill X, Guigó R, de Sanjosé S, Piris MA, Montserrat E, González-Díaz M, Puente XS, Jares P, Valencia A, Himmelbauer H, Quesada V, Bea S, Stratton MR, Futreal PA, Campbell PJ, Vincent-Salomon A, Richardson AL, Reis-Filho JS, van de Vijver M, Thomas G, Masson-Jacquemier JD, Aparicio S, Borg A, Børresen-Dale AL, Caldas C, Foekens JA, Stunnenberg HG, van't Veer L, Easton DF, Spellman PT, Martin S, Barker AD, Chin L, Collins FS, Compton CC, Ferguson ML, Gerhard DS, Getz G, Gunter C, Guttmacher A, Guyer M, Hayes DN, Lander ES, Ozenberger B, Penny R, Peterson J, Sander C, Shaw KM, Speed TP, Spellman PT, Vockley JG, Wheeler DA, Wilson RK, Hudson TJ, Chin L, Knoppers BM, Lander ES, Lichter P, Stein LD, Stratton MR, Anderson W, Barker AD, Bell C, Bobrow M, Burke W, Collins FS, Compton CC, DePinho RA, Easton DF, Futreal PA, Gerhard DS, Green AR, Guyer M, Hamilton SR, Hubbard TJ, Kallioniemi OP, Kennedy KL, Ley TJ, Liu ET, Lu Y, Majumder P, Marra M, Ozenberger B, Peterson J, Schafer AJ, Spellman PT, Stunnenberg HG, Wainwright BJ, Wilson RK, Yang H. International network of cancer genome projects. Nature. 2010 Apr 15; 464(7291):993-998. doi: 10.1038/nature08987. Erratum in: Nature. 2010 Jun 17;465(7300):966. Himmelbaue, Heinz [corrected to Himmelbauer, Heinz]; Gardiner, Brooke A [corrected to Gardiner, Brooke B]; Cross, Anthony [corrected to Cros, Anthony]. [download]
- Kawano S, Shimamura T, Niida A, Imoto S, Yamaguchi R, Nagasaki M, Yoshida R, Print CG, Miyano S. Discovering functional gene pathways associated with cancer heterogeneity via sparse supervised learning. 2010 IEEE International Conference on Bioinformatics and Biomedicine (BIBM). 2010 Dec; 253-258. doi: 10.1109/BIBM.2010.5706572. [download]
- Koh CH, Nagasaki M, Saito A, Wong L, Miyano S. DA 1.0: parameter estimation of biological pathways using data assimilation approach. Bioinformatics. 2010 Jul 15; 26(14):1794-1796. doi: 10.1093/Bioinformatics/btq276. [download]
- Kojima K, Imoto S, Nagasaki M, Miyano S. Gene regulatory network clustering for graph layout based on microarray gene expression data. Genome Informatics. 2010 Jul; 24:84-95. doi: 10.1142/9781848166585_0007. [download]
- Kojima K, Nagasaki M, Miyano S. An efficient biological pathway layout algorithm combining grid-layout and spring embedder for complicated cellular location information. BMC Bioinformatics. 2010 Jun 18; 11:335. doi: 10.1186/1471-2105-11-335. [download]
- Kojima K, Perrier E, Imoto S, Miyano S. Optimal Search on Clustered Structural Constraint for Learning Bayesian Network Structure. J Mach Learn Res. 2010 Mar 1; 11: 285-310. https://dl.acm.org/doi/10.5555/1756006.1756015. [download]
- Kojima K, Yamaguchi R, Imoto S, Yamauchi M, Nagasaki M, Yoshida R, Shimamura T, Ueno K, Higuchi T, Gotoh N, Miyano S. A state space representation of VAR models with sparse learning for dynamic gene networks. Genome Informatics. 2010 Jan; 22:56-68. doi: 10.1142/9781848165786_0006. [download]
- Li C, Nagasaki M, Saito A, Miyano S. Time-dependent structural transformation analysis to high-level Petri net model with active state transition diagram. BMC Syst Biol. 2010 Apr 1; 4:39. doi: 10.1186/1752-0509-4-39. [download]
- Mitou N, Matsuno H, Miyano S, Inouye S. Essential role of Ror gene in the interaction of feedback loops in mammalian circadian clocks. Modeling in Systems Biology – The Petri Net Approach. Koch I, Reisig W, Schreiber F (eds.). Springer. 2010 Oct; 281-306. https://link.springer.com/chapter/10.1007/978-1-84996-474-6_13 [download]
- Miwa Y, Li C, Ge QW, Matsuno H, Miyano S. On determining firing delay time of transitions for Petri net based signaling pathways by introducing stochastic decision rules. In Silico Biol. 2010 Jan 17;10(1):49-66. doi: 10.3233/ISB-2010-0417. [download]
- Miwa Y, Murakami Y, Ge QW, Li C, Matsuno H, Miyano S. Delay Time Determination for the Timed Petri Net Model of a Signaling Pathway Based on Its Structural Information. IEICE Trans Fundam Electron Commun Comput Sci. 2010 Dec 1; 93-A(12): 2717-2729. doi: 10.1587/transfun.E93.A.2717. [download]
- Nagasaki M, Saito A, Jeong E, Li C, Kojima K, Ikeda E, Miyano S. Cell Illustrator 4.0: a computational platform for systems biology. In Silico Biol. 2010 Jan 9;10(1):5-26. doi: 10.3233/ISB-2010-0415. [download]
- Niida A, Imoto S, Nagasaki M, Yamaguchi R, Miyano S. A novel meta-analysis approach of cancer transcriptomes reveals prevailing transcriptional networks in cancer cells. Genome Informatics. 2010 Jan; 22:121-131. doi:10.1142/9781848165786_0010. [download]
- Niida A, Imoto S, Yamaguchi R, Nagasaki M, Fujita A, Shimamura T, Miyano S. Model-free unsupervised gene set screening based on information enrichment in expression profiles. Bioinformatics. 2010 Dec 15;26(24):3090-3097. doi: 10.1093/Bioinformatics/btq592. [download]
- Niida A, Imoto S, Yamaguchi R, Nagasaki M, Miyano S. Gene set-based module discovery decodes cis-regulatory codes governing diverse gene expression across human multiple tissues. PLoS One. 2010 Jun 9; 5(6):e10910. doi: 10.1371/journal.pone.0010910. [download]
- Saito A, Nagasaki M, Miyano S. Hybrid functional Petri net with extension for dynamic pathway modeling. Modeling in Systems Biology – The Petri Net Approach. Koch I, Reisig W, Schreiber F (eds.). Springer. 2010 Oct; 101-120. https://link.springer.com/chapter/10.1007/978-1-84996-474-6_6 [download]
- Sato H, Nakada H, Yamaguchi R, Imoto S, Miyano S, Kami M. When should we intervene to control the 2009 influenza A(H1N1) pandemic? Euro Surveill. 2010 Jan 7; 15(1):19455. doi: 10.2807/ese.15.01.19455-en. [download]
- Shimamura T, Imoto S, Nagasaki M, Yamauchi M, Yamaguchi R, Fujita A, Tamada Y, Gotoh N, Miyano S. Collocation-based sparse estimation for constructing dynamics. Genome Informatics. 2010 Jul; 24:164-178. doi: 10.1142/9781848166585_0014. [download]
- Shimamura T, Imoto S, Niida A, Nagasaki M, Yamaguchi R, Miyano S. Network profiling analysis for generating modulator-dependent gene networks. 2010 IEEE International Conference on Bioinformatics and Biomedicine Workshops (BIBMW). 2010 Dec 18; 1: 836-836. DOI: 10.1109/BIBMW.2010.5703933. [download]
- Shimamura T, Imoto S, Yamaguchi R, Nagasaki M, Miyano S. Inferring dynamic gene networks under varying conditions for transcriptomic network comparison. Bioinformatics. 2010 Apr 15;26(8):1064-1072. doi: 10.1093/Bioinformatics/btq080. [download]
- Tasaki S, Nagasaki M, Kozuka-Hata H, Semba K, Gotoh N, Hattori S, Inoue J, Yamamoto T, Miyano S, Sugano S, Oyama M. Phosphoproteomics-based modeling defines the regulatory mechanism underlying aberrant EGFR signaling. PLoS One. 2010 Nov 10; 5(11):e13926. doi: 10.1371/journal.pone.0013926. [download]
- Yamaguchi R, Imoto S, Miyano S. Network-Based Predictions and Simulations by Biological State Space Models: Search for Drug Mode of Action. J Comput Sci Technol. 2010 Jan; 25(1): 131-153. doi: 10.1007/s11390-010-9311-7. [download]
- Yuji K, Matsumura T, Miyano S, Tsuchiya R, Kami M. Human papillomavirus vaccine coverage. Lancet. 2010 Jul 31; 376(9738):329-330; author reply 330. doi: 10.1016/S0140-6736(10)61180-4. [download]
- Araki H, Tamada Y, Imoto S, Dunmore B, Sanders D, Humphrey S, Nagasaki M, doi: A, Nakanishi Y, Yasuda K, Tomiyasu Y, Tashiro K, Print C, Charnock-Jones DS, Kuhara S, Miyano S. Analysis of PPARalpha-dependent and PPARalpha-independent transcript regulation following fenofibrate treatment of human endothelial cells. Angiogenesis. 2009 Apr 9; 12(3):221-229. doi: 10.1007/s10456-009-9142-8. [download]
- Do JH, Yamaguchi R, Miyano S. Exploring temporal transcription regulation structure of Aspergillus fumigatus in heat shock by state space model. BMC Genomics. 2009 Jul 8; 10: 306. doi: 10.1186/1471-2164-10-306. [download]
- Fujita A, Gomes LR, Sato JR, Yamaguchi R, Thomaz CE, Sogayar MC, Miyano S. Multivariate gene expression analysis reveals functional connectivity changes between normal/tumoral prostates. BMC Syst Biol. 2008 Dec 5; 2: 106. doi: 10.1186/1752-0509-2-106. [download]
- Fujita A, Patriota AG, Sato JR, Miyano S. The impact of measurement errors in the identification of regulatory networks. BMC Bioinformatics. 2009 Dec 13; 10: 412. doi: 10.1186/1471-2105-10-412. [download]
- Fujita A, Sato JR, DA Silva FH, Galvão MC, Sogayar MC, Miyano S. Quality control and reproducibility in DNA microarray experiments. Genome Informatics. 2009 Oct; 23(1):21-31. Download: http://giw.hgc.jp/giw2009/pdf/20-Fujita.pdf [download]
- Fujita A, Sato JR, Demasi MA, Sogayar MC, Ferreira CE, Miyano S. Comparing Pearson, Spearman and Hoeffding's D measure for gene expression association analysis. J Bioinform Comput Biol. 2009 Aug; 7(4):663-684. doi: 10.1142/s0219720009004230. [download]
- Hashimoto TB, Nagasaki M, Kojima K, Miyano S. BFL: a node and edge betweenness based fast layout algorithm for large scale networks. BMC Bioinformatics. 2009 Jan 15; 10: 19. doi: 10.1186/1471-2105-10-19. [download]
- Li C, Nagasaki M, Ueno K, Miyano S. Simulation-based model checking approach to cell fate specification during Caenorhabditis elegans vulval development by hybrid functional Petri net with extension. BMC Syst Biol. 2009 Apr 27; 3: 42. doi: 10.1186/1752-0509-3-42. [download]
- Miyakawa C, Sugii M, Matsuno H, Miyano S. Computational Predictions for Functional Proteins Working after Cleaved in Apoptotic Pathway. 2009 International Conference on Complex, Intelligent and Software Intensive Systems. 2009 Mar; 807-812. doi: 10.1109/CISIS.2009.197. [download]
- Miyano S, Yamaguchi R, Tamada Y, Nagasaki M, Imoto S. Gene Networks Viewed through Two Models. Lecture Notes in Bioinformatics. 2009 Apr; 5462: 54-66. doi: 10.1007/978-3-642-00727-9_8. [download]
- Nakamura K, Yoshida R, Nagasaki M, Miyano S, Higuchi T. Parameter estimation of in silico biological pathways with particle filtering towards a petascale computing. Pac Symp Biocomput. 2009 Jan; 14: 227-238. https://psb.stanford.edu/psb-online/proceedings/psb09/nakamura.pdf [download]
- Shimamura T, Imoto S, Yamaguchi R, Fujita A, Nagasaki M, Miyano S. Recursive regularization for inferring gene networks from time-course gene expression profiles. BMC Syst Biol. 2009 Apr 22; 3: 41. doi: 10.1186/1752-0509-3-41. [download]
- Tamada Y, Araki H, Imoto S, Nagasaki M, doi: A, Nakanishi Y, Tomiyasu Y, Yasuda K, Dunmore B, Sanders D, Humphreys S, Print C, Charnock-Jones DS, Tashiro K, Kuhara S, Miyano S. Unraveling dynamic activities of autocrine pathways that control drug-response transcriptome networks. Pac Symp Biocomput. 2009 Jan; 14: 251-263. https://psb.stanford.edu/psb-online/proceedings/psb09/tamada.pdf [download]
- Watanabe-Fukuda Y, Yamamoto M, Miura N, Fukutake M, Ishige A, Yamaguchi R, Nagasaki M, Saito A, Imoto S, Miyano S, Takeda J, Watanabe K. Orengedokuto and berberine improve indomethacin-induced small intestinal injury via adenosine. J Gastroenterol. 2009 Mar 25; 44(5):380-389. doi: 10.1007/s00535-009-0005-2. [download]
- Yamamoto T, Bannai H, Nagasaki M, Miyano S. Better Decomposition Heuristics for the Maximum-Weight Connected Graph Problem Using Betweenness Centrality. Lecture Notes in Artificial Intelligence. 2009 Oct; 5808: 465-472. doi: 10.1007/978-3-642-04747-3_40. [download]
- Yashiro Y, Bannai H, Minowa T, Yabiku T, Miyano S, Osawa M, Iwama A, Nakauchi H. Transcriptional profiling of hematopoietic stem cells by high-throughput sequencing. Int J Hematol. 2009 Jan 20; 89(1):24-33. doi: 10.1007/s12185-008-0212-2. [download]
- Yoshikawa N, Nagasaki M, Sano M, Tokudome S, Ueno K, Shimizu N, Imoto S, Miyano S, Suematsu M, Fukuda K, Morimoto C, Tanaka H. Ligand-based gene expression profiling reveals novel roles of glucocorticoid receptor in cardiac metabolism. Am J Physiol Endocrinol Metab. 2009 Jun; 296(6):E1363-1373. doi: 10.1152/ajpendo.90767.2008. [download]
- Do JH, Miyano S. The GC and window-averaged DNA curvature profile of secondary metabolite gene cluster in Aspergillus fumigatus genome. Appl Microbiol Biotechnol. 2008 Oct; 80(5):841-847. doi: 10.1007/s00253-008-1638-4. [download]
- Fujita A, Sato JR, Garay-Malpartida HM, Sogayar MC, Ferreira CE, Miyano S. Modeling nonlinear gene regulatory networks from time series gene expression data. J Bioinform Comput Biol. 2008 Oct; 6(5):961-979. doi: 10.1142/s0219720008003746. [download]
- Hatanaka Y, Nagasaki M, Yamaguchi R, Obayashi T, Numata K, Fujita A, Shimamura T, Tamada Y, Imoto S, Kinoshita K, Nakai K, Miyano S. A novel strategy to search conserved transcription factor binding sites among coexpressing genes in human. Genome Informatics. 2008 Oct; 20: 212-221. doi: 10.11234/gi1990.20.212. [download]
- Hioka K, Miwa Y, Li C, Ge QW, Matsuno H, Miyano S. An algorithm to estimate delay times in petri net models of signaling pathways with experimental data. BIONETICS '08: Proceedings of the 3rd International Conference on Bio-Inspired Models of Network, Information and Computing Systems. 2008 Nov 25; 1: 1–8. https://dl.acm.org/doi/10.5555/1512504.1512506. [download]
- Hirose O, Yoshida R, Imoto S, Yamaguchi R, Higuchi T, Charnock-Jones DS, Print C, Miyano S. Statistical inference of transcriptional module-based gene networks from time course gene expression profiles by using state space models. Bioinformatics. 2008 Apr 1; 24(7):932-942. doi: 10.1093/Bioinformatics/btm639. [download]
- Hirose O, Yoshida R, Yamaguchi R, Imoto S, Higuchi T, Miyano S. Analyzing Time Course Gene Expression Data with Biological and Technical Replicates to Estimate Gene Networks by State Space Models. 2008 Second Asia International Conference on Modelling & Simulation (AMS). 2008 May; 940-946. doi: 10.1109/AMS.2008.150. [download]
- Jeong E, Nagasaki M, Miyano S. Rule-based reasoning for system dynamics in cell systems. Genome Informatics. 2008 Oct; 20: 25-36. doi: 10.11234/gi1990.20.25. [download]
- Kojima K, Fujita A, Shimamura T, Imoto S, Miyano S. Estimation of nonlinear gene regulatory networks via L1 regularized NVAR from time series gene expression data. Genome Informatics. 2008 Oct; 20: 37-51. doi: 10.11234/gi1990.20.37. [download]
- Kojima K, Nagasaki M, Miyano S. Fast grid layout algorithm for biological networks with sweep calculation. Bioinformatics. 2008 Jun 15; 24(12):1433-1441. doi: 10.1093/Bioinformatics/btn196. [download]
- Mitou N, Ikegami Y, Matsuno H, Miyano S, Inouye ST. Simulation analysis for the effect of light-dark cycle on the entrainment in circadian rhythm. Genome Informatics. 2008 Nov 7; 21:212-223. doi: 10.11234/gi1990.21.212. [download]
- Nagasaki M, Saito A, Li C, Jeong E, Miyano S. Systematic reconstruction of TRANSPATH data into cell system markup language. BMC Syst Biol. 2008 Jun 23; 2: 53. doi: 10.1186/1752-0509-2-53. [download]
- Numata K, Imoto S, Miyano S. Partial Order-Based Bayesian Network Learning Algorithm for Estimating Gene Networks. 2008 IEEE International Conference on Bioinformatics and Biomedicine. 2008 Nov 03-05; 357-360. doi: 10.1109/BIBM.2008.85. [download]
- Numata K, Yoshida R, Nagasaki M, Saito A, Imoto S, Miyano S. ExonMiner: Web service for analysis of GeneChip Exon array data. BMC Bioinformatics. 2008 Nov 26; 9: 494. doi: 10.1186/1471-2105-9-494. [download]
- Yamaguchi R, Imoto S, Yamauchi M, Nagasaki M, Yoshida R, Shimamura T, Hatanaka Y, Ueno K, Higuchi T, Gotoh N, Miyano S. Predicting differences in gene regulatory systems by state space models. Genome Informatics. 2008 Nov 7; 21: 101-113. doi: 10.11234/gi1990.21.101. [download]
- Yoshida R, Nagasaki M, Yamaguchi R, Imoto S, Miyano S, Higuchi T. Bayesian learning of biological pathways on genomic data assimilation. Bioinformatics. 2008 Nov 15; 24(22):2592-2601. doi: 10.1093/Bioinformatics/btn483. [download]
- Affara M, Dunmore B, Savoie C, Imoto S, Tamada Y, Araki H, Charnock-Jones DS, Miyano S, Print C. Understanding endothelial cell apoptosis: what can the transcriptome, glycome and proteome reveal? Philos Trans R Soc Lond B Biol Sci. 2007 Aug 29; 362(1484):1469-1487. doi: 10.1098/rstb.2007.2129. [download]
- Akutsu T, Bannai H, Miyano S, Ott S. On the complexity of deriving position specific score matrices from positive and negative sequences. Discret Appl Math. 2007 Apr; 155(6-7): 676-685. doi: 10.1016/j.dam.2004.10.011. [download]
- Fujita A, Sato JR, Garay-Malpartida HM, Yamaguchi R, Miyano S, Sogayar MC, Ferreira CE. Modeling gene expression regulatory networks with the sparse vector autoregressive model. BMC Syst Biol. 2007 Aug 30; 1:1 39. doi: 10.1186/1752-0509-1-39. [download]
- Gupta PK, Yoshida R, Imoto S, Yamaguchi R, Miyano S. Statistical Absolute Evaluation of Gene Ontology Terms with Gene Expression Data. Lecture Notes in Bioinformatics. 2007 May; 4463: 146-157. doi: 10.1007/978-3-540-72031-7_14. [download]
- Hirose O, Yoshida R, Yamaguchi R, Imoto S, Higuchi T, Miyano S. Clustering samples characterized by time course gene expression profiles using the mixture of state space models. Genome Informatics. 2007 Dec; 18: 258-266. doi: 10.11234/gi1990.18.258. [download]
- Imoto S, Miyano S. Bayesian network approach to estimate gene networks. A Mittal A, Kassim A, Tan T (Eds.). Bayesian Network Technologies: Applications and Graphical Models. Idea Group Publishers, USA. 2007 Mar 31; 269-299. https://www.igi-global.com/gateway/chapter/5505 https://doi.org/10.4018/978-1-59904-141-4.ch013 [download] [download]
- Imoto S, Tamada Y, Savoie CJ, Miyano S. Analysis of gene networks for drug target discovery and validation. Methods Mol Biol. 2007; 360: 33-56. doi: 10.1385/1-59745-165-7:33. [download]
- Jeong E, Nagasaki M, Miyano S. Conversion from BioPAX to CSO for system dynamics and visualization of biological pathway. Genome Informatics. 2007 Dec; 18: 225-236. doi: 10.11234/gi1990.18.225. [download]
- Jeong E, Nagasaki M, Saito A, Miyano S. Cell system ontology: representation for modeling, visualizing, and simulating biological pathways. In Silico Biol. 2007 Oct; 7(6):623-638. https://content.iospress.com/articles/in-silico-biology/isb00335. [download]
- Numata K, Imoto S, Miyano S. A Structure Learning Algorithm for Inference of Gene Networks from Microarray Gene Expression Data Using Bayesian Networks. 2007 IEEE 7th International Symposium on BioInformatics and BioEngineering. 2007 Oct; 1280-1284. doi: 10.1109/BIBE.2007.4375731. [download]
- Kojima K, Nagasaki M, Jeong E, Kato M, Miyano S. An efficient grid layout algorithm for biological networks utilizing various biological attributes. BMC Bioinformatics. 2007 Mar 6; 8: 76. doi: 10.1186/1471-2105-8-76. [download]
- Li C, Ge QW, Nakata M, Matsuno H, Miyano S. Modelling and simulation of signal transductions in an apoptosis pathway by using timed Petri nets. J Biosci. 2007 Jan;32(1):113-127. doi: 10.1007/s12038-007-0011-6. Erratum in: J Biosci. 2007 Jun; 32(4):805. [download]
- Saito A, Nagasaki M, Oyama M, Kozuka-Hata H, Semba K, Sugano S, Yamamoto T, Miyano S. AYUMS: an algorithm for completely automatic quantitation based on LC-MS/MS proteome data and its application to the analysis of signal transduction. BMC Bioinformatics. 2007 Jan 18; 8: 15. doi: 10.1186/1471-2105-8-15. [download]
- Shimamura T, Imoto S, Yamaguchi R, Miyano S. Weighted lasso in graphical Gaussian modeling for large gene network estimation based on microarray data. Genome Informatics. 2007 Dec; 19: 142-153. doi: 10.11234/gi1990.19.142. [download]
- Sugii M, Okada R, Matsuno H, Miyano S. Performance improvement in protein N-myristoyl classification by BONSAI with insignificant indexing symbol. Genome Informatics. 2007 Dec; 18: 277-286. doi: 10.11234/gi1990.18.277. [download]
- Yamaguchi R, Yamamoto M, Imoto S, Nagasaki M, Yoshida R, Tsuiji K, Ishige A, Asou H, Watanabe K, Miyano S. Identification of activated transcription factors from microarray gene expression data of Kampo medicine-treated mice. Genome Informatics. 2007 Dec; 18: 119-129. doi: 10.11234/gi1990.18.119. [download]
- Yamaguchi R, Yoshida R, Imoto S, Higuchi T, Miyano S. Finding module-based gene networks with state-space models - Mining high-dimensional and short time-course gene expression data. IEEE Signal Process Mag. 2007 Jan; 24(1): 37-46. doi: 10.1109/MSP.2007.273053. [download]
- Yoshida R, Numata K, Imoto S, Nagasaki M, Doi A, Ueno K, Miyano S. Computational Genome-Wide Discovery of Aberrant Splice Variations with Exon Expression Profiles. 2007 IEEE 7th International Symposium on BioInformatics and BioEngineering. 2007 Oct; 715-722. doi: 10.1109/BIBE.2007.4375639. [download]
- Doi A, Nagasaki M, Matsuno H, Miyano S. Simulation-based validation of the p53 transcriptional activity with hybrid functional petri net. In Silico Biol. 2006 Jan 4; 6(1-2):1-13. https://content.iospress.com/articles/in-silico-biology/isb00216 [download]
- Doi A, Nagasaki M, Ueno K, Matsuno H, Miyano S. A combined pathway to simulate CDK-dependent phosphorylation and ARF-dependent stabilization for p53 transcriptional activity. Genome Informatics. 2006 Aug; 17(1):112-123. Doi: 10.11234/gi1990.17.112. [download]
- Imoto S, Tamada Y, Araki H, Yasuda K, Print CG, Charnock-Jones SD, Sanders D, Savoie CJ, Tashiro K, Kuhara S, Miyano S. Computational strategy for discovering druggable gene networks from genome-wide RNA expression profiles. Pac Symp Biocomput. 2006 Jan; 11: 559-571. https://psb.stanford.edu/psb-online/proceedings/psb06/imoto.pdf [download]
- Jeong E, Miyano S. A Weighted Profile Based Method for Protein-RNA Interacting Residue Prediction. Lecture Notes in Computer Science. 2006 Apr 6; 3939: 123-139. doi: 10.1007/11732488_11. [download]
- Li C, Suzuki S, Ge QW, Nakata M, Matsuno H, Miyano S. Structural modeling and analysis of signaling pathways based on Petri nets. J Bioinform Comput Biol. 2006 Oct; 4(5):1119-1140. doi: 10.1142/s021972000600234x. [download]
- Matsuno H, Inouye ST, Okitsu Y, Fujii Y, Miyano S. A new regulatory interaction suggested by simulations for circadian genetic control mechanism in mammals. J Bioinform Comput Biol. 2006 Feb; 4(1):139-153. doi: 10.1142/s021972000600176x. [download]
- Nagasaki M, Yamaguchi R, Yoshida R, Imoto S, doi: A, Tamada Y, Matsuno H, Miyano S, Higuchi T. Genomic data assimilation for estimating hybrid functional Petri net from time-course gene expression data. Genome Informatics. 2006 Aug; 17(1):46-61. doi: 10.11234/gi1990.17.46. [download]
- Okada R, Sugii M, Matsuno H, Miyano S. Machine Learning Prediction of Amino Acid Patterns in Protein N-myristoylation. Lecture Notes in Bioinformatics. 2006 Aug; 4146: 4-14. doi: 10.1007/11818564_2. [download]
- Saito A, Nagasaki M, doi: A, Ueno K, Miyano S. Cell fate simulation model of gustatory neurons with MicroRNAs double-negative feedback loop by hybrid functional Petri net with extension. Genome Informatics. 2006 Aug; 17(1):100-111. doi: 10.11234/gi1990.17.100. [download]
- Takei Y, Kawakoshi A, Tsukada T, Yuge S, Ogoshi M, Inoue K, Hyodo S, Bannai H, Miyano S. Contribution of comparative fish studies to general endocrinology: structure and function of some osmoregulatory hormones. J Exp Zool A Comp Exp Biol. 2006 Sep 1; 305(9):787-798. doi: 10.1002/jez.a.309. [download]
- Tasaki S, Nagasaki M, Oyama M, Hata H, Ueno K, Yoshida R, Higuchi T, Sugano S, Miyano S. Modeling and estimation of dynamic EGFR pathway by data assimilation approach using time series proteomic data. Genome Informatics. 2006 Dec; 17(2):226-238. doi: 10.11234/gi1990.17.2_226. [download]
- Yoshida R, Higuchi T, Imoto S, Miyano S. ArrayCluster: an analytic tool for clustering, data visualization and module finder on gene expression profiles. Bioinformatics. 2006 Jun 15; 22(12):1538-1539. doi: 10.1093/Bioinformatics/btl129. [download]
- Yoshida R, Numata K, Imoto S, Nagasaki M, doi: A, Ueno K, Miyano S. A statistical framework for genome-wide discovery of biomarker splice variations with GeneChip Human Exon 1.0 ST Arrays. Genome Informatics. 2006 Aug; 17(1):88-99. doi: 10.11234/gi1990.17.88. [download]
- De Hoon MJ, Makita Y, Nakai K, Miyano S. Prediction of transcriptional terminators in Bacillus subtilis and related species. PLoS Comput Biol. 2005 Aug; 1(3):e25. doi: 10.1371/journal.pcbi.0010025. [download]
- Hirose O, Nariai N, Tamada Y, Bannai H, Imoto S, Miyano S. Estimating Gene Networks from Expression Data and Binding Location Data via Boolean Networks. Lecture Notes in Computer Science. 2005 Apr 27; 3482: 349-356. doi: 10.1007/11424857_38. [download]
- Kitakaze H, Matsuno H, Ikeda N, Miyano S. Prediction of debacle points for robustness of biological pathways by using recurrent neural networks. Genome Informatics. 2005 Aug; 16(1):192-202. doi: 10.11234/gi1990.16.192. [download]]
- Makita Y, de Hoon MJ, Ogasawara N, Miyano S, Nakai K. Bayesian joint prediction of associated transcription factors in Bacillus subtilis. Pac Symp Biocomput. 2005 Jan; 10: 507-518. doi: 10.1142/9789812702456_0048. http://psb.stanford.edu/psb-online/proceedings/psb05/makita.pdf [download]
- Nagasaki M, Doi: A, Matsuno H, Miyano S. Computational modeling of biological processes with Petri net-based architecture. Bioinformatics Technologies (Chen YP (ed)). Springer Press. 2005 Jan 18; 179-243. https://link.springer.com/chapter/10.1007/3-540-26888-X_7 DOI: https://doi.org/10.1007/3-540-26888-X_7 [download]
- Nariai N, Tamada Y, Imoto S, Miyano S. Estimating gene regulatory networks and protein-protein interactions of Saccharomyces cerevisiae from multiple genome-wide data. Bioinformatics. 2005 Sep 1; 21 Suppl 2:ii206-12. doi: 10.1093/Bioinformatics/bti1133. [download]
- Ohtsubo S, Iida A, Nitta K, Tanaka T, Yamada R, Ohnishi Y, Maeda S, Tsunoda T, Takei T, Obara W, Akiyama F, Ito K, Honda K, Uchida K, Tsuchiya K, Yumura W, Ujiie T, Nagane Y, Miyano S, Suzuki Y, Narita I, Gejyo F, Fujioka T, Nihei H, Nakamura Y. Association of a single-nucleotide polymorphism in the immunoglobulin mu-binding protein 2 gene with immunoglobulin A nephropathy. J Hum Genet. 2005 Jan 1; 50(1):30-35. doi: 10.1007/s10038-004-0214-8. [download]
- Ott S, Hansen A, Kim SY, Miyano S. Superiority of network motifs over optimal networks and an application to the revelation of gene network evolution. Bioinformatics. 2005 Jan 15; 21(2):227-238. doi: 10.1093/Bioinformatics/bth484. [download]
- Tamada Y, Bannai H, Imoto S, Katayama T, Kanehisa M, Miyano S. Utilizing evolutionary information and gene expression data for estimating gene networks with bayesian network models. J Bioinform Comput Biol. 2005 Dec; 3(6):1295-1313. doi: 10.1142/s0219720005001569. [download]
- Tamada Y, Imoto S, Tashiro K, Kuhara S, Miyano S. Identifying drug active pathways from gene networks estimated by gene expression data. Genome Informatics. 2005 Aug; 16(1):182-191. doi: 10.11234/gi1990.16.182. [download]
- Ando T, Imoto S, Miyano S. Functional Data Analysis of the Dynamics of Gene Regulatory Networks. Lecture Notes in Artificial Intelligence. 2004 Nov; 3303: 69-83. doi: 10.1007/978-3-540-30478-4_7. [download]
- Ando T, Imoto S, Miyano S. Kernel mixture survival models for identifying cancer subtypes, predicting patient's cancer types and survival probabilities. Genome Informatics. 2004 Dec; 15(2):201-210. doi: 10.11234/gi1990.15.2_201. [download]
- Bannai H, Hyyrö H, Shinohara A, Takeda M, Nakai K, Miyano S. An O(N2) algorithm for discovering optimal Boolean pattern pairs. IEEE/ACM Trans Comput Biol Bioinform. 2004 Oct-Dec; 1(4):159-170. doi: 10.1109/TCBB.2004.36. [download]
- Bannai H, Hyyrö H, Shinohara A, Takeda M, Nakai K, Miyano S. Finding Optimal Pairs of Patterns. Lecture Notes in Bioinformatics. 2004 Sep; 3240: 450-462. doi: 10.1007/978-3-540-30219-3_38. [download]
- Bannai H, Inenaga S, Shinohara A, Takeda M, Miyano S. Efficiently finding regulatory elements using correlation with gene expression. J Bioinform Comput Biol. 2004 Jun; 2(2):273-288. doi: 10.1142/s0219720004000612. [download]
- De Hoon MJ, Imoto S, Kobayashi K, Ogasawara N, Miyano S. Predicting the operon structure of Bacillus subtilis using operon length, intergene distance, and gene expression information. Pac Symp Biocomput. 2004 Jan; 9: 276-287. doi: 10.1142/9789812704856_0027. https://psb.stanford.edu/psb-online/proceedings/psb04/dehoon.pdf. [download]
- De Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004 Jun 12; 20(9):1453-1454. doi: 10.1093/Bioinformatics/bth078. [download]
- De Hoon MJ, Makita Y, Imoto S, Kobayashi K, Ogasawara N, Nakai K, Miyano S. Predicting gene regulation by sigma factors in Bacillus subtilis from genome-wide data. Bioinformatics. 2004 Aug 4; 20 Suppl 1:i101-8. doi: 10.1093/Bioinformatics/bth927. [download]
- Doi A, Fujita S, Matsuno H, Nagasaki M, Miyano S. Constructing biological pathway models with hybrid functional Petri nets. In Silico Biol. 2004 Apr 7; 4(3):271-91. https://content.iospress.com/articles/in-silico-biology/isb00133. [download]
- Imoto S, Higuchi T, Goto T, Tashiro K, Kuhara S, Miyano S. Combining microarrays and biological knowledge for estimating gene networks via Bayesian networks. J Bioinform Comput Biol. 2004 Mar; 2(1):77-98. doi: 10.1142/s021972000400048x. [download]
- Imoto S, Higuchi T, Kim SY, Jeong E, Miyano S. Residual Bootstrapping and Median Filtering for Robust Estimation of Gene Networks from Microarray Data. Lecture Notes in Bioinformatics. 2004 May; 3082:149-160. doi: 10.1007/978-3-540-25974-9_12. [download]
- Inenaga S, Bannai H, Hyyrö H, Shinohara A, Takeda M, Nakai K, Miyano S. Finding Optimal Pairs of Cooperative and Competing Patterns with Bounded Distance. Lecture Notes in Artificial Intelligence. 2004 Oct; 3245: 32-46. doi: 10.1007/978-3-540-30214-8_3. [download]
- Jeong E, Chung IF, Miyano S. A neural network method for identification of RNA-interacting residues in protein. Genome Informatics. 2004 Oct; 15(1):105-116. doi: 10.11234/gi1990.15.105. [download]
- Kim S, Imoto S, Miyano S. Dynamic Bayesian network and nonparametric regression for nonlinear modeling of gene networks from time series gene expression data. Biosystems. 2004 Jul; 75(1-3):57-65. doi: 10.1016/j.Biosystems.2004.03.004. [download]
- Matsui M, Fujita S, Suzuki S, Matsuno H, Miyano S. Simulated Cell Division Processes of the Xenopus Cell Cycle Pathway by Genomic Object Net. J Integr Bioinform. 2004 Dec 20; 1(1) 3. doi: 10.1515/jib-2004-3. [download]
- Nagasaki M, Doi A, Matsuno H, Miyano S. Integrating Biopathway Databases for Large-scale Modeling and Simulation. Proc. Second Asia-Pacific Bioinformatics Conference (APBC 2004). 2004 Jan; 43-52. https://dl.acm.org/doi/abs/10.5555/976520.976527. [download]
- Nagasaki M, doi: A, Matsuno H, Miyano S. A versatile petri net based architecture for modeling and simulation of complex biological processes. Genome Informatics. 2004 Oct; 15(1):180-197. doi: 10.11234/gi1990.15.180. [download]
- Nakamichi R, Imoto S, Miyano S. Case-Control Study of Binary Disease Trait Considering Interactions between SNPs and Environmental Effects using Logistic Regression. Proc. 4th IEEE International Symposium on BioInformatics and BioEngineering (BEBE 2004). IEEE Computer Society. 2004 May; 73-78. doi: 10.1109/BIBE.2004.1317327. [download]
- Nariai N, Kim S, Imoto S, Miyano S. Using protein-protein interactions for refining gene networks estimated from microarray data by Bayesian networks. Pac Symp Biocomput. 2004 Jan; 9: 336-347. doi: 10.1142/9789812704856_0032. https://psb.stanford.edu/psb-online/proceedings/psb04/nariai.pdf. [download]
- Ott S, Imoto S, Miyano S. Finding optimal models for small gene networks. Pac Symp Biocomput. 2004 Jan; 9: 557-567. doi: 10.1142/9789812704856_0052. http://psb.stanford.edu/psb-online/proceedings/psb04/ott.pdf. [download]
- Takei Y, Inoue K, Ogoshi M, Kawahara T, Bannai H, Miyano S. Identification of novel adrenomedullin in mammals: a potent cardiovascular and renal regulator. FEBS Lett. 2004 Jan 2; 556(1-3):53-58. doi: 10.1016/s0014-5793(03)01368-1. [download]
- Miyano S. Computational systems biology. Proceedings of the Third International Conference on Information (Info’04). 2004 Dec; 9-14. [download]
- Ando T, Imoto S, Miyano S. Bayesian network and radial basis function network regression for nonlinear modeling of genetic network. Proceedings of the Third International Conference on Information (Info’04). 2004 Dec; 561-564. [download]
- Nakano M, Noda R, Kitakaze H, Matsuno H, Miyano S. XML pathway file conversion between Genomic Object Net and SBML. Proceedings of the Third International Conference on Information (Info’04). 2004 Dec; 585-588. [download]
- De Hoon MJ, Imoto S, Kobayashi K, Ogasawara N, Miyano S. Inferring gene regulatory networks from time-ordered gene expression data of Bacillus subtilis using differential equations. Pac Symp Biocomput. 2003 Jan; 8: 17-28. doi: 10.1142/9789812776303_0003. https://psb.stanford.edu/psb-online/proceedings/psb03/dehoon.pdf. [download]
- Doi A, Nagasaki M, Fujita S, Matsuno H, Miyano S. Genomic Object Net: II. Modelling biopathways by hybrid functional Petri net with extension. Appl Bioinformatics. 2003; 2(3):185-188. Download: https://www.semanticscholar.org/paper/Genomic-Object-Net%3A-II.-Modelling-biopathways-by-doi:-Nagasaki/8473d90fffcf4f206b92757b19779dab78bc3ee9 [download]
- Imoto S, Higuchi T, Goto T, Tashiro K, Kuhara S, Miyano S. Combining Microarrays and Biological Knowledge for Estimating Gene Networks via Bayesian Networks. Proceedings of the 2003 IEEE Bioinformatics Conference (CSB2003). 2003 Aug 11-14; 1: 104-113. DOI: 10.1109/CSB.2003.1227309. https://www.computer.org/csdl/proceedings-article/csb/2003/20000104/12OmNz4SOvM [download] (Extended Abstract) Complete Version: J Bioinform Comput Biol. 2003 Jul; 1(2):231-52)
- Imoto S, Kim S, Goto T, Miyano S, Aburatani S, Tashiro K, Kuhara S. Bayesian network and nonparametric heteroscedastic regression for nonlinear modeling of genetic network. J Bioinform Comput Biol. 2003 Jul; 1(2):231-52. doi: 10.1142/s0219720003000071. [download]
- Imoto S, Savoie CJ, Aburatani S, Kim S, Tashiro K, Kuhara S, Miyano S. Use of gene networks for identifying and validating drug targets. J Bioinform Comput Biol. 2003 Oct; 1(3):459-74. doi: 10.1142/s0219720003000290. [download]
- Kim SY, Imoto S, Miyano S. Inferring gene networks from time series microarray data using dynamic Bayesian networks. Brief Bioinform. 2003 Sep; 4(3):228-235. doi: 10.1093/bib/4.3.228. [download]
- Matsuno H, Fujita S, Doi A, Nagasaki M, Miyano S. Towards Biopathway Modeling and Simulation. Lecture Notes in Computer Science. 2003 May; 2679: 3-22. doi: 10.1007/3-540-44919-1_2. [download]
- Matsuno H, Murakami R, Yamane R, Yamasaki N, Fujita S, Yoshimori H, Miyano S. Boundary formation by notch signaling in Drosophila multicellular systems: experimental observations and gene network modeling by Genomic Object Net. Pac Symp Biocomput. 2003 Jan; 8: 152-63. doi: 10.1142/9789812776303_0015. http://psb.stanford.edu/psb-online/proceedings/psb03/matsuno.pdf. [download]
- Matsuno H, Tanaka Y, Aoshima H, doi: A, Matsui M, Miyano S. Biopathways representation and simulation on hybrid functional Petri net. In Silico Biol. 2003 Jun 23; 3(3):389-404. https://content.iospress.com/articles/in-silico-biology/isb00103. [download]
- Miyano S. Inference, Modeling and Simulation of Gene Networks. Lecture Notes in Computer Science. 2003 Feb; 2602:207-211. doi: 10.1007/3-540-36481-1_31. [download]
- Nagasaki M, Doi A, Matsuno H, Miyano S. Recreating Biopathway Databases towards Simulation. Lecture Notes in Computer Science. 2003 Feb; 2602: 168-169. doi: 10.1007/3-540-36481-1_18. [download]
- Nagasaki M, doi: A, Matsuno H, Miyano S. Genomic Object Net: I. A platform for modelling and simulating biopathways. Appl Bioinformatics. 2003; 2(3):181-184. https://www.semanticscholar.org/paper/Genomic-Object-Net%3A-I.-A-platform-for-modelling-and-Nagasaki-doi:/faf51f92bbde8f860a25c6a021bcc7bb3649cc83. [download]
- Obara W, Iida A, Suzuki Y, Tanaka T, Akiyama F, Maeda S, Ohnishi Y, Yamada R, Tsunoda T, Takei T, Ito K, Honda K, Uchida K, Tsuchiya K, Yumura W, Ujiie T, Nagane Y, Nitta K, Miyano S, Narita I, Gejyo F, Nihei H, Fujioka T, Nakamura Y. Association of single-nucleotide polymorphisms in the polymeric immunoglobulin receptor gene with immunoglobulin A nephropathy (IgAN) in Japanese patients. J Hum Genet. 2003 May 23; 48(6):293-299. doi: 10.1007/s10038-003-0027-1. [download]
- Ott S, Miyano S. Finding optimal gene networks using biological constraints. Genome Informatics. 2003 Dec; 14: 124-133. doi: 10.11234/gi1990.14.124. [download]
- Ott S, Tamada Y, Bannai H, Nakai K, Miyano S. Intrasplicing - analysis of long intron sequences. Pac Symp Biocomput. 2003 Jan; 8: 339-50. doi: 10.1142/9789812776303_0032. https://psb.stanford.edu/psb-online/proceedings/psb03/ott.pdf. [download]
- Savoie CJ, Aburatani S, Watanabe S, Eguchi Y, Muta S, Imoto S, Miyano S, Kuhara S, Tashiro K. Use of gene networks from full genome microarray libraries to identify functionally relevant drug-affected genes and gene regulation cascades. DNA Res. 2003 Feb 28;10(1):19-25. doi: 10.1093/dnares/10.1.19. Erratum in: DNA Res. 2003 Apr 30;10(2): following pg. 84. [download]
- Tamada Y, Kim S, Bannai H, Imoto S, Tashiro K, Kuhara S, Miyano S. Estimating gene networks from gene expression data by combining Bayesian network model with promoter element detection. Bioinformatics. 2003 Oct; 19 Suppl 2:ii227-236. doi: 10.1093/Bioinformatics/btg1082. [download]
- Akutsu T, Kuhara S, Maruyama O, Miyano S. Identification of genetic networks by strategic gene disruptions and gene overexpressions under a boolean model. Theoret Comput Sci. 2003 April 4; 298(1): 235-251. Doi: 10.1016/S0304-3975(02)00425-5.] [download]
- Akiyama F, Tanaka T, Yamada R, Ohnishi Y, Tsunoda T, Maeda S, Takei T, Obara W, Ito K, Honda K, Uchida K, Tsuchiya K, Nitta K, Yumura W, Nihei H, Ujiie T, Nagane Y, Miyano S, Suzuki Y, Fujioka T, Narita I, Gejyo F, Nakamura Y. Single-nucleotide polymorphisms in the class II region of the major histocompatibility complex in Japanese patients with immunoglobulin A nephropathy. J Hum Genet. 2002 Oct 1; 47(10):532-8. doi: 10.1007/s100380200080. [download]
- Akutsu T, Bannai H, Miyano S, Ott O. On the Complexity of Deriving Position Specific Score Matrices from Examples. Lecture Notes in Computer Science. 2002 Jun 19; 2373: 168-177. doi: 10.1007/3-540-45452-7_15. [download]
- Akutsu T, Miyano S. Selecting informative genes for cancer classification using gene expression data. Computational and Statistical Approaches to Genomics. Zhang W, Shmulevich I (eds.) Kluwer Academic Pub. 2002 Jan 1; 79-92. DOI: 10.1007/0-387-26288-1_6 https://link.springer.com/chapter/10.1007/0-387-26288-1_6 [download]
- Bannai H, Inenaga S, Shinohara A, Takeda M, Miyano S. A string pattern regression algorithm and its application to pattern discovery in long introns. Genome Informatics. 2002 Dec; 13: 3-11. doi: 10.11234/gi1990.13.3. [download]
- Bannai H, Tamada Y, Maruyama O, Nakai K, Miyano S. Extensive feature detection of N-terminal protein sorting signals. Bioinformatics. 2002 Feb; 18(2):298-305. doi: 10.1093/Bioinformatics/18.2.298. [download]
- De Hoon MJ, Imoto S, Miyano S. Inferring Gene Regulatory Networks from Time-Ordered Gene Expression Data Using Differential Equations. Lecture Notes in Computer Science. 2002 Nov; 2534: 267-274. doi: 10.1007/3-540-36182-0_24. [download]
- De Hoon MJ, Imoto S, Miyano S. Statistical analysis of a small set of time-ordered gene expression data using linear splines. Bioinformatics. 2002 Nov; 18(11):1477-1485. doi: 10.1093/Bioinformatics/18.11.1477. [download]
- Imoto S, Goto T, Miyano S. Estimation of genetic networks and functional structures between genes by using Bayesian networks and nonparametric regression. Pac Symp Biocomput. 2002 Jan; 7: 175-186. doi: 10.1142/9789812799623_0017. https://psb.stanford.edu/psb-online/proceedings/psb02/imoto.pdf. [download]
- Imoto S, Kim S, Goto T, Aburatani S, Tashiro K, Kuhara S, Miyano S. Bayesian network and nonparametric heteroscedastic regression for nonlinear modeling of genetic network. IEEE Computer Society Bioinformatics Conference (CSB2002). 2002 Aug; 1: 219-227. doi: 10.1109/CSB.2002.1039344. http://citeseerx.ist.psu.edu/viewdoc/download?doi:=10.1.1.411.9558&rep=rep1&type=pdf. [download]
- Maruyama O, Bannai H, Tamada Y, Kuhara S, Miyano S. Fast algorithm for extracting multiple unordered short motifs using bit operations. Information Sciences. 2002 Oct; 146(1-4): 115-126. doi: 10.1016/S0020-0255(02)00219-0. [download]
- Maruyama O, Shoudai T, Miyano S. Toward Drawing an Atlas of Hypothesis Classes: Approximating a Hypothesis via Another Hypothesis Model. Lecture Notes in Computer Science. 2002 Nov; 2373: 220-232. doi: 10.1007/3-540-36182-0_20. [download]
- Takei T, Iida A, Nitta K, Tanaka T, Ohnishi Y, Yamada R, Maeda S, Tsunoda T, Takeoka S, Ito K, Honda K, Uchida K, Tsuchiya K, Suzuki Y, Fujioka T, Ujiie T, Nagane Y, Miyano S, Narita I, Gejyo F, Nihei H, Nakamura Y. Association between single-nucleotide polymorphisms in selectin genes and immunoglobulin A nephropathy. Am J Hum Genet. 2002 Mar; 70(3):781-786. doi: 10.1086/339077. [download]
- Tamada Y, Bannai H, Maruyama O, Miyano S. Foundations of Designing Computational Knowledge Discovery Processes. Lecture Notes in Artificial Intelligence. 2002 Mar; 2281: 459-470. doi: 10.1007/3-540-45884-0_34. [download-]
- Bannai H, Tamada Y, Maruyama O, Miyano S. VML: A View Modeling Language for Computational Knowledge Discovery. Lecture Notes in Artificial Intelligence. 2001 Dec; 2226: 30-44. doi: 10.1007/3-540-45650-3_6. [download]
- Bannai H, Tamada Y, Maruyama O, Nakai K, Miyano S. Views: Fundamental Building Blocks in the Process of Knowledge Discovery. Proceedings of the Fourteenth International Florida Artificial Intelligence Research Society Conference. AAAI Press. 2001 May 21: 233-238. https://dl.acm.org/doi/10.5555/646814.708164. https://www.aaai.org/Papers/FLAIRS/2001/FLAIRS01-045.pdf . [download]
- Maruyama O, Shoudai T, Furuichi E, Kuhara S, Miyano S. Learning Conformation Rules. Lecture Notes in Artificial Intelligence. 2001 Dec; 2226: 243-257. doi: 10.1007/3-540-45650-3_22. [download]
- Matsuno H, doi: A, Hirata Y, Miyano S. XML documentation of biopathways and their simulations in Genomic Object Net. Genome Informatics. 2001 Dec; 12: 54-62. doi: 10.11234/gi1990.12.54. [download]
- Sim KL, Uchida T, Miyano S. ProDDO: a database of disordered proteins from the Protein Data Bank (PDB). Bioinformatics. 2001 Apr;17(4):379-380. doi: 10.1093/Bioinformatics/17.4.379. [download]
- Akutsu T, Miyano S, Kuhara S. A simple greedy algorithm for finding functional relations: efficient implementation and average case analysis. Lecture Notes in Artificial Intelligence. 2000 Dec; 1967: 86-98. doi: 10.1007/3-540-44418-1_8. [download]
- Akutsu T, Miyano S, Kuhara S. Algorithms for identifying Boolean networks and related biological networks based on matrix multiplication and fingerprint function. J Comput Biol. 2000 Aug; 7(3-4): 331-343. doi: 10.1089/106652700750050817. [download]
- Akutsu T, Miyano S, Kuhara S. Algorithms for inferring qualitative models of biological networks. Pac Symp Biocomput. 2000 Jan; 5: 293-304. doi: 10.1142/9789814447331_0028. https://psb.stanford.edu/psb-online/proceedings/psb00/akutsu.pdf. [download]
- Akutsu T, Miyano S, Kuhara S. Inferring qualitative relations in genetic networks and metabolic pathways. Bioinformatics. 2000 Aug; 16(8):727-34. doi: 10.1093/Bioinformatics/16.8.727. [download]
- Maruyama O, Miyano S. Design Aspects of Discovery Systems. IEICE Trans. Inf. and Sys. 2000 Jan 25; E83-D(1): 61-70. http://citeseerx.ist.psu.edu/viewdoc/download?doi:=10.1.1.29.8291&rep=rep1&type=pdf [download]
- Matsuno H, doi: A, Nagasaki M, Miyano S. Hybrid Petri net representation of gene regulatory network. Pac Symp Biocomput. 2000 Jan; 5: 341-352. doi: 10.1142/9789814447331_0032. https://psb.stanford.edu/psb-online/proceedings/psb00/matsuno.pdf. [download]
- Miyano S., Shinohara A, Shinohara T: Polynomial-time learning of elementary formal systems. New Generation Computing. 2000 Sep; 18: 217-242. doi: 10.1007/BF03037530. [download]
- Tanaka M, Nakazono S, Matsuno H, Tsujimoto H, Kitamura Y, Miyano S. Intelligent system for topic survery in MEDLINE by keyword recommendation and learning text characteristics. Genome Informatics. 2000 Dec; 11: 73-82. doi: 10.11234/gi1990.11.73. [download]
- Akutsu T, Miyano S, Kuhara S. Identification of genetic networks from a small number of gene expression patterns under the boolean network model. Pac Symp Biocomput. 1999 Jan; 4: 17-28. doi: 10.1142/9789814447300_0003. https://psb.stanford.edu/psb-online/proceedings/psb99/Akutsu.pdf. [download]
- Akutsu T, Miyano S. On the approximation of protein threading. Theoretical Computer Science. 1999 Jan 17; 21(2):261-275. doi: 10.1016/S0304-3975(98)00089-9. [download]
- Bannai H, Miyano S. A definition of discovery in terms of generalized descriptional complexity. Lecture Notes in Artificial Intelligence. 1999 Oct; 1721: 316-318. doi: 10.1007/3-540-46846-3_29. [download]
- Maruyama O, Uchida T, K.L. Sim, Miyano S. Designing views in HypothesisCreator: System for assisting in discovery. Lecture Notes in Artificial Intelligence. 1999 Oct; 1721: 115-127. doi: 10.1007/3-540-46846-3_11. [download]
- Moriyama O, Shinohara A, Takeda M, Maruyama O, Goto T, Miyano S, Kuhara S. A system to find genetic networks using weighted network model. Genome Informatics. 1999 Dec; 10: 186-195. doi: 10.11234/gi1990.10.186. [download]
- Nagasaki M, Onami S, Miyano S, Kitano H. Bio-calculus: its concept and molecular interaction. Genome Informatics. 1999 Dec; 10: 133-143. doi: 10.11234/gi1990.10.133. [download]
- Yasuda T, Bannai H, Onami S, Miyano S, Kitano H. Towards automatic construction of cell-lineage of C. elegans from Normarski DIC microscope images. Genome Informatics. 1999 Dec; 10: 144-154. doi: 10.11234/gi1990.10.144. [download]
- Akutsu T, Maruyama O, Miyano S, Kuhara S. A system for identifying genetic networks from gene expression patterns produced by gene disruptions and overexpressions. Genome Informatics. 1998 Dec; 9: 151-160. doi: 10.11234/gi1990.9.151. [download]
- Maruyama O, Uchida Shoudai T, Miyano S. Toward Genomic Hypothesis Creator: View Designer for Discovery. Lecture Notes in Artificial Intelligence. 1998 Dec; 1532: 105-116. doi: 10.1007/3-540-49292-5_10. [download]
- Noda K, Shinohara A, Takeda M, S. Matsumoto, Miyano S, Kuhara S. Finding genetic network from experiments by weighted network model. Genome Informatics. 1998 Dec; 9: 141-150. doi: 10.11234/gi1990.9.141. [download]
- Usuzaka S, K.L. Sim, Tanaka M, Matsuno H, Miyano S. A machine learning approach to reducing the work of experts in article selection from database: a case study for regulatory relations of S. cerevisiae genes in MEDLINE. Genome Informatics. 1998 Dec; 9: 91-101. doi: 10.11234/gi1990.9.91. [download]
- Akutsu T, Kuhara S, Maruyama O, Miyano S. Identification of gene regulatory networks by strategic gene disruptions and gene overexpressions. Proc. 9th Annual ACM SIAM Symposium on Discrete Algorithms (SODA’98). 1998 Jan 1: 695-702. https://dl.acm.org/doi/10.5555/314613.315050. [download]
- Yamaguchi A, Nakano K, Miyano S. An approximation algorithm for the minimum common supertree problem. Nordic J. Computing. 1997 Sep 1; 4(3): 303-316. [download]
- Furukawa N, Matsumoto S, Shinohara A, Shoudai T, Miyano S. HAKKE: A multi-strategy prediction system for sequences. Genome Informatics. 1996 Nov; 7: 98-107. doi: 10.11234/gi1990.7.98. [download]
- Maruyama O, Miyano S. Inferring a tree from walks. Theoretical Computer Science. 1996 Jul 15; 161(1-2): 289-300. doi: 10.1016/0304-3975(95)00156-5. [download]
- Maruyama O, Miyano S. Taking a walk on a graph. Mathematica Japonica. 1996 May. 43(3): 595-606. https://www.semanticscholar.org/paper/TAKING-A-WALK-ON-A-GRAPH-Maruyama-Miyano/2a9f5f612c4e11712e34ea583a55450dd5ca4039. [download]
- Miyano S. Genome Informatics: New Frontiers of Computer Science and Biosciences. Proc. International Symposium on Cooperative Database Systems for Advanced Applications. Advanced Database Research and Development Series. Kambayashi Y, Yokota K (eds). 1996 Dec; 7: 12-21. https://www.worldscientific.com/worldscibooks/10.1142/3496. Review [download]
- Miyano S, Shimozono S, Maruyama O. Some algorithmic problems arising from genome informatics. Advances in Computing Techniques - Algorithms, Databases and Parallel Processing. Imai H, Wong WF, Loe KF (eds). World Scientific. 1996 Mar; 45-59. https://www.worldscientific.com/worldscibooks/10.1142/2969#t=aboutBook Review [download]
- Tateishi E, Maruyama O, Miyano S. Extracting best consensus motifs from positive and negative examples. Lecture Notes in Computer Science. 1996 Feb; 1046: 219-230. doi: 10.1007/3-540-60922-9_19. [download]
- Tateishi E, Miyano S. A greedy strategy for finding motifs from yes-no examples. A greedy strategy for finding motifs from yes-no examples. Pac Symp Biocomput. 1996 Jan; 1: 599-613. PMID: 9390261. https://psb.stanford.edu/psb-online/proceedings/psb99/Akutsu.pdf. [download]
- Maruyama O, Miyano S. Graph inference from a walk for trees of bounded degree 3 is NP-complete. Lecture Notes in Computer Science. 1995 Aug; 969: 257-266. doi: 10.1007/3-540-60246-1_132. [download]
- Miyano S. Learning theory towards genome informatics. IEICE Transactions on Information and Systems. 1995 May 25; E78-D(5): 560-567. Review. https://search.ieice.org/bin/summary.php?id=e78-d_5_560. doi: 10.1007/3-540-57370-4_34. Springer Link [download]
- Shimozono S, Miyano S. Complexity of finding alphabet indexing. IEICE Transactions on Information and Systems. 1995 Jan 25; E78-D(1): 13-18. doi: https://search.ieice.org/bin/summary.php?id=e78-d_1_13. [download]
- Shoudai T, Lappe M, Miyano S, Shinohara A, Okazaki T, Arikawa S, Uchida T, Shimozono S, Shinohara T, Kuhara S: BONSAI Garden: parallel knowledge discovery system for amino acid sequences. Proceedings of the Third International Conference on Intelligent Systems for Molecular Biology. 1995 Aug; AAAI Press. 3: 359-366. https://www.aaai.org/Papers/ISMB/1995/ISMB95-043.pdf. [download]
- Shoudai T, Miyano S. Using maximal independent sets to solve problems in parallel. Theoretical Computer Science. 1995 Aug; 148(1): 57-65. doi: 10.1016/0304-3975(94)00221-4. [download]
- Uchida T, Shoudai T, Miyano S. Parallel algorithms for refutation tree problem on elementary formal graph systems. IEICE Transactions on Information and Systems. 1995 Feb 25. E78-D(2): 99-112. https://search.ieice.org/bin/summary.php?id=e78-d_2_99. [download]
- Shimozono S, Shinohara A, Shinohara T, Miyano S, Kuhara S and Arikawa S. Knowledge acquisition from amino acid sequences by machine learning system BONSAI. Transactions on Information Processing Society of Japan. 1994 Oct 15; 35(10): 2009-2018. JSPS Best Paper Award. doi: https://ipsj.ixsq.nii.ac.jp/ej/index.php?active_action=repository_view_main_item_detail&page_id=13&block_id=8&item_id=14104&item_no=1. [download]
- Uchida T, Shoudai T, Miyano S. Polynomial time algorithm solving the refutation tree problem for formal graph systems. Bulletin of Informatics and Cybernetics. 1994 Mar; 26(1/2): 55-74. doi: 10.5109/3069. [download]
- Arikawa S, Kuhara S, Miyano S, Y. Mukouchi, Shinohara A, Shinohara T. A machine discovery from amino acid sequences by decision trees over regular patterns. New Generation Computing. 1993 Sep; 11: 361-375. doi: 10.11234/gi1990.4.74. [download]
- Arikawa S, Shinohara T, Miyano S, Shinohara A. More About Learning Elementary Formal Systems. Lecture Notes in Artificial Intelligence. 1993 Feb 26. 659: 107-117. doi: 10.1007/BFb0030388. [download]
- Furuya S, Miyano S. NP-hard aspects in analogical reasoning. Bulletin of lnformatics and Cybernetics. 1993 Mar; 25(3/4): 155-159. doi: 10.5109/13428. [download]
- Shimozono S, Shinohara A, Shinohara T, Miyano S, Kuhara S, Arikawa S. Finding alphabet indexing for decision trees over regular patterns: an approach to bioinformatical knowledge acquisition. Proceedings of the Twenty-Sixth Hawaii International Conference on System Sciences. IEEE Computer Society. 1993 Jan; 763-772. doi: 10.1109/HICSS.1993.270664. [download]
- Shinohara A, Shimozono S, Uchida T, Miyano S, Kuhara S, Arikawa S. Running learning systems in parallel for machine discovery from sequences. Genome Informatics. 1993 Dec; 4: 74-83. doi: 10.11234/gi1990.4.74. [download]
- Shoudai T, Miyano S. A parallel algorithm for the maximal co-hitting set problem. IEICE Transactions on Information and Systems. 1993 Feb 25; E76-D (2): 296-298. https://search.ieice.org/bin/summary.php?id=e76-d_2_296&category=D&year=1993&lang=E&abst=. [download]
- Uchida T, Miyano S. O(log*n) time parallel algorithm for computing bounded degree maximal subgraphs. Journal of Information Processing. 1993 Mar 31; 16(1): 16-20. https://ipsj.ixsq.nii.ac.jp/ej/?action=pages_view_main&active_action=repository_view_main_item_detail&item_id=59596&item_no=1&page_id=13&block_id=8 [download]
- Arikawa S, Kuhara S, Miyano S, Shinohara A, Shinohara T. A learning algorithm for elementary formal systems and its experiments on identification of transmembrane domains. Proceedings of the Twenty-Fifth Hawaii International Conference on System Sciences. 1992 Jan; Vol. I, IEEE Computer Society. 675-684. doi: 10.1109/HICSS.1992.183220. [download]
- Arikawa S, Miyano S, Shinohara A, Shinohara T, Yamamoto A. Algorithmic learning theory with elementary formal systems. IEICE Transactions on Information and Systems. 1992 Jul; E75-D(4): 405-414. doi: https://search.ieice.org/bin/summary.php?id=e75-d_4_405. [download]
- Furuya S, Miyano S. NP-complete problems on label updating calculation in ATMS. Bulletin of Informatics and Cybernetics. 1992 Mar 27; 25(1/2): 1-5. doi: 10.5109/3146. [download]
- Arikawa S, Miyano S, Shinohara A, Shimozono S, Shinohara T, Kuhara S. Knowledge acquisition from amino acid sequences by learning algorithms. Proc. 2nd Japanese Knowledge Acquisition for Knowledge-Based Systems Workshop (JKAW ’92). Nov 1992; 109-128. Invited Paper. [download]
- Furuya S, Miyano S. Analogy is NP-hard. Proceedings of the Second Workshop on Algorithmic Learning Theory. IOS Press. 1991 Oct; 207-212. https://www.google.co.jp/books/edition/Algorithmic_Learning_Theory_II/UNsV29DK-L8C?hl=ja&gbpv=1&dq=Analogy+is+NP-hard&pg=PA207&printsec=frontcover [download]
- Miyano S, Shinohara A, Shinohara T. Which classes of elementary formal systems are polynomial-time learnable? Proceedings of the Second Workshop on Algorithmic Learning Theory. ISO Press. 1991 Oct; 139-150. https://books.google.co.jp/books?id=UNsV29DK-L8C&pg=PA139&lpg=PA139&dq=Which+classes+of+elementary+formal+systems+are+polynomial-time+learnable&source=bl&ots=MpwtV2XnUM&sig=ACfU3U1NXqxSc7J65u-HNyujEHpVJjsQbg&hl=ja&sa=X&ved=2ahUKEwjDtZS52crxAhVCyYsBHbukBisQ6AEwCHoECAkQAw#v=onepage&q=Which%20classes%20of%20elementary%20formal%20systems%20are%20polynomial-time%20learnable&f=false. [download]
- Miyano S. Δp2-complete lexicographically maximal subgraph problems. Theoretical Computer Science. 1991 Sep 30; 88(1): 33-57. doi: 10.1016/0304-3975(91)90072-A. [download]
- Shinohara A, Miyano S. Teachability in computational learning. New Generation Computing. 1991 Feb; 8(4): 333-347. doi: 10.1007/BF03037091. [download]
- Miyano S. Systematized approaches to the complexity of subgraph problems. Journal of Information Processing. 1990 Feb 10; 13(4): 442-448. Invited Review. https://ipsj.ixsq.nii.ac.jp/ej/?action=pages_view_main&active_action=repository_view_main_item_detail&item_id=59722&item_no=1&page_id=13&block_id=8. [download]
- Miyano S, Shiraishi S, Shoudai T. A List of P-complete Problems. RIFIS Technical Report 17, Kyushu University Institutional Repository. 1990 Dec 29 Revised; 1989 Oct 11 First Version;1-69. https://catalog.lib.kyushu-u.ac.jp/opac_download_md/3123/rifis-tr-17.pdf Review [download]
- Shoudai T, Miyano S. Bounded degree maximal subgraph problems are in NC. Proc Toyohashi Symposium on Theoretical Computer Science. 1990 Aug; 97-101. https://catalog.lib.kyushu-u.ac.jp/opac_detail_md/?reqCode=frombib&lang=0&amode=MD100000&opkey=B161996754180642&bibid=3134&start=1 [download]
- Arikawa S, Haraguchi M, Inoue H, Kawasaki Y, Miyahara T, Miyano S, Oshima K, Sakai H, Shinohara T, Shiraishi S,Takeda M, Takeya S, Yamamoto A, Yuasa H. The text database management system SIGMA: an improvement of the main engine. Proceedings of Berliner lnformatik-Tage. 1989 Jun. 72-81. [download]
- Miyano S. The lexicographically first maximal subgraph problems - P-completeness and NC algorithms. Mathematical Systems Theory. 1989 Dec. 22(1): 47-73. doi: 10.1007/BF02088292. [download]
- Miyano S. A parallelizable lexicographically first edge-induced subgraph problem. Information Processing Letters. 1988 Feb 29; 27(2): 75-78. doi: 10.1016/0020-0190(88)90095-6. [download]
- Miyano S. Indexing alternating finite automata and binary tree like circuits, Bulletin of lnformatics and Cybernetics. 1988 Mar; 23(1/2): 79-88. doi: 10.5109/13396. [download]
- Miyano S. Parallel complexity and P-complete problems. Proceedings of International Conference on Fifth Generation Computer Systems. 1988 Nov-Dec; 532-541. https://www.ueda.info.waseda.ac.jp/AITEC_ICOT_ARCHIVES/ICOT/Museum/FGCS/FGCS88en-proc2/88eMPCT-1.pdf. [download]
- Miyano S. The complexity of the lexicographically first maximal rooted tree problem. Fachbereich Mathematik-Informatik, Universität-Gesamthochschule-Paderborn. 1987 Jul; Nr. 42. [download]
- Hayashi T, Miyano S. Finite tree automata on infinite trees. Bulletin of Informatics and Cybernetics. 1985 Mar; 21(3/4): 71-82. doi: 10.5109/13369. [download]
- Miyano S, Hayashi T. Alternating finite automata on ω-words. Theoretical Computer Science. 1984 Jun 21; 32(3): 321-330. doi: 10.1016/0304-3975(84)90049-5. Preliminary version: CAAP 1984: 195-210. [download]
- Miyano S. Remarks on two-way automata with weak-counters. Information Processing Letters. 1984 Feb 28; 18(2): 105-107. doi: 10.1016/0020-0190(84)90032-2. [download]
- Miyano S. Remarks on multihead pushdown automata and multihead stack automata. Journal of Computer and System Sciences. 1983 Aug; 27(1): 116-124. doi: 10.1016/0022-0000(83)90032-6. [download]
- Miyano S, Haraguchi M. Recovery of incomplete tables under functional dependencies. Bulletin of Informatics and Cybernetics. 1982 Mar; 20: (1/2): 25-41. doi: 10.5109/13334. [download]
- Miyano S. A hierarchy theorem for multihead stack-counter automata. Acta Informatica. 1982 Apr; 17: 63–67. doi: 10.1007/BF00262976. [download]
- Miyano S. Two-way deterministic multi-weak-counter machines. Theoretical Computer Science. 1982 Oct; 21(1): 27-37. doi: 10.1016/0304-3975(82)90086-X. [download]
- Hayashi T, Miyano S. Flow expressions and complexity analysis. IPSJ Technical Reports (PRO). Information Processing Society of Japan. 1982 Oct 18; 35(1982-PRO-002): 1-10. https://ipsj.ixsq.nii.ac.jp/ej/index.php?active_action=repository_view_main_item_detail&page_id=13&block_id=8&item_id=31478&item_no=1 [download]
- Miyano S. One-way weak-stack-counter automata. Journal of Computer and System Sciences. 1980 Feb; 20(1): 59-76. doi: 10.1016/0022-0000(80)90005-7. [download]
- Miyano S. ON A LOWER BOUND OF SHEPHERDSON FUNCTION. Memoirs of the Faculty of Science, Kyushu University. Series A, Mathematics. 1979 Mar; 33 (2): 257-267. doi: 10.2206/kyushumfs.33.257. [download]
- Miyano S. A note on one-way pushdown automata with weak-counters. Research Report No. 94, Research Institute of Fundamental Information Science, Kyushu University. 1979 Aug; pp.13. [download]
- Hirokawa S, Miyano S. A NOTE ON THE REGURALITY OF FUZZY LANGUAGES. Memoirs of the Faculty of Science, Kyushu University. Series A, Mathematics. 1978 Oct; 32 (1): 61-66. doi: 10.2206/kyushumfs.32.61. [download]]
- Miyano S. ON AN AUTOMATON WHICH RECOGNIZES A FAMILY OF AUTOMATA. Memoirs of the Faculty of Science, Kyushu University. Series A, Mathematics. 1978 Oct; 32(1): 37-51. doi: 10.2206/kyushumfs.32.37. [download]
- Ma B, Rajasekaran S, Huan J, Miyano S, Shehu A, Gombar VK, Schapranow M, Yoo I, Zhou J, Hu XT (Eds). Proceedings of 2015 IEEE International Conference on Bioinformatics and Biomedicine (BIBM). 9-12 November 2015, Washington, DC, USA. IEEE Computer Society Press. https://www.computer.org/csdl/proceedings/bibm/2015/12OmNyYm2xh https://ieeexplore.ieee.org/xpl/conhome/7350074/proceeding
- Chen XW, Miyano S (Eds). Special Issue on “The First IEEE Conference on Healthcare Informatics, Imaging, and Systems Biology HISB'11”. J Bioinform Comput Biol. 2011 Oct; 9(5): (Preface: v-vi. doi: 10.1142/s0219720011005719). PMID: 22069831 https://www.worldscientific.com/doi/abs/10.1142/S0219720011005719 [download]
- Mandoiu II, Miyano S, Przytycka TM, Rajasekaran S (Eds). Proceedings of the IEEE International Conference on Computational Advances in Bio and Medical Sciences (ICCABS). 3-5 February 2011, Orlando, FL, USA. IEEE Computer Society Press. DOI: 10.1109/ICCABS18686.2011. https://ieeexplore.ieee.org/xpl/conhome/5724107/proceeding
- Ferreira CE, Miyano S, Stadler PF. Advances in Bioinformatics and Computational Biology. 5th Brazilian Symposium on Bioinformatics, BSB 2010, Rio de Janeiro, Brazil, August 31-September 3, 2010, Proceedings. Lecture Notes in Bioinformatics. 2010 Aug; 6268. https://link.springer.com/book/10.1007/978-3-642-15060-9
- Akutsu T, Kanehisa M, Klipp E, Miyano S, Mohr S, Tullius T, Wallach I (Eds). Genome Informatics 2010. Genome Informatics Series Vol. 24. Proceedings of the 10th Annual International Workshop on Bioinformatics and Systems Biology (IBSB 2010), Kyoto University, Japan, 26 – 28 July 2010. https://doi.org/10.1142/p756 | July 2010. Pages: 244. https://www.worldscientific.com/worldscibooks/10.1142/p756#t=aboutBook
- DeLisi C, Kanehisa M, Miyano S, Mohr S, Wallach I (Eds). Genome Informatics 2009. Genome Informatics Series Vol. 22. Proceedings of the 9th Annual International Workshop on Bioinformatics and Systems Biology (IBSB 2009), Boston University, Boston, USA, 27 – 29 July 2009. https://doi.org/10.1142/p718 | January 2010. Pages: 228. https://www.worldscientific.com/worldscibooks/10.1142/p718#t=toc
- Knapp E-W, Benson G, Holzhütter H-G, Kanehisa M, Miyano S (Eds). Genome Informatics 2008. Genome Informatics Series Vol. 20. Proceedings of The 8th International Workshop on Bioinformatics and Systems Biology (IBSB 2008), Teikyo Hotel, Zeuten Lake, Germany, 9-10 June 2008. October 2008. Imperial College Press. Pages: 288. https://play.google.com/books/reader?id=suAxEAAAQBAJ&pg=GBS.PR3&hl=ja https://www.jstage.jst.go.jp/browse/gi/20/0/_contents/-char/en [download]
- Brazma A, Miyano S, Akutsu T (Eds). Proceedings of the 6th Asia-Pacific Bioinformatics Conference. Series on Advances in Bioinformatics and Computational Biology: Volume 6. Kyoto, Japan, 14 – 17 January 2008. https://doi.org/10.1142/p544 | December 2007. Pages: 412. https://www.worldscientific.com/worldscibooks/10.1142/p544#t=aboutBook
- Miyano S, DeLisi C, Holzhütter H-G, Kanehisa M (Eds). Genome Informatics 2007. Genome Informatics Series Vol. 18. Proceedings of the 7th Annual International Workshop on Bioinformatics and Systems Biology (IBSB 2007), Institute of Medical Science, University of Tokyo, Japan, 31 July – 2 August 2007. https://doi.org/10.1142/p540 | December 2007. Pages: 348. https://www.worldscientific.com/worldscibooks/10.1142/p540#t=aboutBook
- DeLisi C, Kanehisa M, Heinrich R, Miyano S (Eds). Genome Informatics 2006. Genome Informatics Series Vol. 17(1). Proceedings of the 6th Annual International Workshop on Bioinformatics and Systems Biology (IBSB 2006), Boston University, USA, 24-26 July 2006. Pages: 251. Universal Academy Press, Tokyo. https://iss.ndl.go.jp/books/R100000002-I000008369023-00. https://www.jstage.jst.go.jp/browse/gi/17/1/_contents/-char/en
- Sakakibara Y, Smith TF, Kanehisa M, Miyano S, Takagi T (Eds). Genome Informatics 2006. Genome Informatics Series Vol. 17(2). Proceedings of the 17th International Conference on Genome Informatics (GIW 2006), Yokohama, Japan, 18-20 December 2006. Pages: 299. Universal Academy Press, Tokyo. https://iss.ndl.go.jp/books/R100000002-I000008503846-00. https://www.jstage.jst.go.jp/browse/gi/17/0/_contents/-char/en
- Miyano S, Mesirov J, Kasif S, Istrail S, Pevzner PA, Waterman M. Research in Computational Molecular Biology. 9th Annual International Conference, RECOMB 2005, Cambridge, MA, USA, May 14-18, 2005, Proceedings. Lecture Notes in Bioinformatics. 2005 Apr 28; 3500. https://link.springer.com/book/10.1007/b135594
- Heinrich T, DeLisi C, Kanehisa M, Miyano S (Eds). Genome Informatics 2005. Genome Informatics Series Vol. 16(1). Proceedings of the 5th Annual International Workshop on Bioinformatics and Systems Biology (IBSB 2005), Humboldt University, Berlin, Germany, 22-25 August 2005. Universal Academy Press, Tokyo. Pages: 281. Universal Academy Press, Tokyo. https://iss.ndl.go.jp/books/R100000002-I000007893689-00 https://www.jstage.jst.go.jp/browse/gi/16/1/_contents/-char/en
- Heinrich R, Mamitsuka H, Kanehisa M, Miyano S, Takagi T (Eds). Genome Informatics 2005. Genome Informatics Series Vol. 15(2). Proceedings of the 16th International Conference on Genome Informatics (GIW 2005), Yokohama, Japan, 19-21 December 2005. Pages: 293. Universal Academy Press, Tokyo. Pages: 305. https://iss.ndl.go.jp/books/R100000002-I000007893686-00 https://www.jstage.jst.go.jp/browse/gi/16/2/_contents/-char/en
- Mamitsuka H, Smith TF, Holzhütter H-G, Kanehisa M, DeLisi C, Heinrich R, Miyano S (Eds). Genome Informatics 2004. Genome Informatics Series Vol. 15(1). Proceedings of the 5th Annual International Workshop on Bioinformatics and Systems Biology (IBSB 2005), Kyoto University, Uji, Japan, 31 May - 3 June 2004. Universal Academy Press, Tokyo. Pages: 281. Universal Academy Press, Tokyo. https://iss.ndl.go.jp/books/R100000002-I000007487072-00 https://www.jstage.jst.go.jp/browse/gi/15/1/_contents/-char/en
- Akutsu T, Brusic V, Miyano S, Takagi T, Kanehisa M (Eds). Genome Informatics 2004. Genome Informatics Series Vol. 15(2). Proceedings of the 15th International Conference on Genome Informatics (GIW 2004), Yokohama, Japan, 13-15 December 2004. Pages: 293. Universal Academy Press, Tokyo. https://iss.ndl.go.jp/books/R100000002-I000007487072-00 https://www.jstage.jst.go.jp/browse/gi/15/2/_contents/-char/en
- Mamitsuka H, Smith TF, Holzhütter H-G, Kanehisa M, DeLisi C, Heinrich R, Miyano S (Eds). Genome Informatics 2004. Genome Informatics Series Vol. 15(1). Proceedings of the 5th Annual International Workshop on Bioinformatics and Systems Biology (IBSB 2005), Kyoto University, Uji, Japan, 31 May - 3 June 2004. Universal Academy Press, Tokyo. Pages: 281. Universal Academy Press, Tokyo. https://iss.ndl.go.jp/books/R100000002-I000007487072-00 https://www.jstage.jst.go.jp/browse/gi/15/1/_contents/-char/en [download]
- Akutsu T, Brusic V, Miyano S, Takagi T, Kanehisa M (Eds). Genome Informatics 2004. Genome Informatics Series Vol. 15(2). Proceedings of the 15th International Conference on Genome Informatics (GIW 2004), Yokohama, Japan, 13-15 December 2004. Pages: 293. Universal Academy Press, Tokyo. https://iss.ndl.go.jp/books/R100000002-I000007487072-00 https://www.jstage.jst.go.jp/browse/gi/15/2/_contents/-char/en
- Gribskov M, Kanehisa M, Miyano S, Takagi T (Eds). Genome Informatics 2003. Genome Informatics Series Vol. 14. Proceedings of the 14th International Conference on Genome Informatics (GIW 2003), Yokohama, Japan, 14-17 December 2003. Pages: 733. Universal Academy Press, Tokyo. https://iss.ndl.go.jp/books/R100000002-I000004364855-00 https://www.jstage.jst.go.jp/browse/gi/14/0/_contents/-char/en
- Lathrop R, Nakai K, Miyano S, Takagi T, Kanehisa M (Eds). Genome Informatics 2002. Genome Informatics Series Vol. 13. Proceedings of the 13th International Conference on Genome Informatics (GIW 2002), Yebisu Garden Place, Tokyo, Japan, 16-18 December 2002. Pages: 603. Universal Academy Press, Tokyo. https://iss.ndl.go.jp/books/R100000002-I000004010804-00 https://www.jstage.jst.go.jp/browse/gi/13/0/_contents/-char/en
- Matsuda H, Miyano S, Takagi T, Wong L (Eds). Genome Informatics 2001. Genome Informatics Series Vol. 12. Proceedings of the 12th International Conference on Genome Informatics (GIW 2001), Yebisu Garden Place, Tokyo, Japan, 17-19 December 2001. Pages: 520. Universal Academy Press, Tokyo. https://iss.ndl.go.jp/books/R100000002-I000003586473-00 https://www.jstage.jst.go.jp/browse/gi/12/0/_contents/-char/en
- Miyano S (Ed.). Special Issue on Surveys on Discovery Science. IEICE Transactions on Information Systems. 2000 Jan 25; 83-D(1). https://globals.ieice.org/en_transactions/information/E83-D_1
- Dunker AK, Konagaya A, Miyano S, Takagi T (Eds). Genome Informatics 2000. Genome Informatics Series Vol. 11. Proceedings of the 11th International Conference on Genome Informatics (GIW 2000), Yebisu Garden Place, Tokyo, Japan,18-19 December 2000. Pages: 472. Universal Academy Press, Tokyo. https://iss.ndl.go.jp/books/R100000002-I000003086133-00 https://www.jstage.jst.go.jp/browse/gi/11/0/_contents/-char/en
- Miyano S, Shamir R, Takagi T (Eds). Currents in Computational Molecular Biology. Frontier Science Series No. 30. January 2000. Universal Academy Press. Pages: 257.
- Shamir R, Miyano S, Istrail S, Pevzner P, Waterman M (Eds). Proceedings of the Fourth Annual International Conference on Computational Molecular Biology (RECOBM 2000). 8-11 April 2000, Tokyo Big Sight, Japan. Pages: 329. https://dl.acm.org/doi/proceedings/10.1145/332306
- Miyano S (Ed.). Special Issue on “Genome Informatics”. Theoretical Computer Science. 1999 Jan 17; 210(2). https://www.sciencedirect.com/journal/theoretical-computer-science/vol/210/issue/2
- Asai K, Miyano S, Takagi T (Eds). Genome Informatics 1999. Genome Informatics Series Vol. 10. Proceedings of the 10th International Conference on Genome Informatics (GIW 1999), Yebisu Garden Place, Tokyo, Japan,14-15 December 1999. Pages: 373. Universal Academy Press, Tokyo. https://iss.ndl.go.jp/books/R100000002-I000003086132-00 https://www.jstage.jst.go.jp/browse/gi/10/0/_contents/-char/en
- Miyano S, Takagi T (Eds). Genome Informatics 1998. Genome Informatics Series Vol. 9. Proceedings of the 9th International Conference on Genome Informatics (GIW 1998), Yebisu Garden Place, Tokyo, Japan,10-11 December 1998. Pages: 400. Universal Academy Press, Tokyo. https://iss.ndl.go.jp/books/R100000002-I000008184631-00 https://www.jstage.jst.go.jp/browse/gi/9/0/_contents/-char/en
- Miyano S, Takagi T (Eds). Genome Informatics 1997. Genome Informatics Series Vol. 8. Proceedings of the 8th International Conference on Genome Informatics (GIW 1997), Yebisu Garden Place, Tokyo, Japan,12-13 December 1997. Pages: 365. Universal Academy Press, Tokyo. https://iss.ndl.go.jp/books/R100000002-I000008179221-00 https://www.jstage.jst.go.jp/browse/gi/8/0/_contents/-char/en
- Asano T, Igarashi Y, Nagamochi H, Miyano S, Suri S. Algorithms and Computation. 7th International Symposium, ISAAC '96, Osaka, Japan, December 16-18, 1996, Proceedings. Lecture Notes in Computer Science. 1996 Dec; 1178. https://link.springer.com/book/10.1007/BFb0009475
- Akutsu T, Asai K, Hagiya M, Kuhara S, Miyano S, Nakai K (Eds). Genome Informatics 1996. Genome Informatics Series Vol. 7. Proceedings of the 7th International Conference on Genome Informatics (GIW 1996), Yebisu Garden Place, Tokyo, Japan, 2-3 December 1996. Pages: 284. Universal Academy Press, Tokyo. https://iss.ndl.go.jp/books/R100000002-I000008179214-00 https://www.jstage.jst.go.jp/browse/gi/7/0/_contents/-char/en
- Hagiya M, Miyano S, Nakai K, Suyama A, Yokomori T, Takagi T (Eds). Genome Informatics 1995. Genome Informatics Series Vol. 6. Proceedings of Genome Informatics Workshop 1995 (GIW 1995), Yokohama, Japan, 11-12 December 1995. Pages: 179. Universal Academy Press, Tokyo. https://iss.ndl.go.jp/books/R100000002-I000008179203-00 https://www.jstage.jst.go.jp/browse/gi/6/0/_contents/-char/en
- Miyano S, Akutsu T, Imai H, Gotoh O, Takagi T (Eds). Genome Informatics 1994. Genome Informatics Series Vol. 5. Proceedings of Genome Informatics Workshop 1994 (GIW 1994), Yokohama, Japan, 19-24 December 1994. Pages: 243. Universal Academy Press, Tokyo. https://iss.ndl.go.jp/books/R100000002-I000002513154-00 https://www.jstage.jst.go.jp/browse/gi/5/0/_contents/-char/en
- Takagi T, Imai H, Miyano S, Mitaku S, Kanehisa M (Eds). Genome Informatics 1993. Genome Informatics Series Vol. 4. Proceedings of Genome Informatics Workshop IV (GIW 1993), Yokohama, Japan, 13-15 December 1993. Pages: 465. Universal Academy Press, Tokyo. https://iss.ndl.go.jp/books/R100000002-I000002352384-00 https://www.jstage.jst.go.jp/browse/gi/4/0/_contents/-char/en
- Satoru Miyano. Digesting Cancer Big Networks by Explainable AI and Supercomputers. The 3rd R-CCS International Symposium (R-CCSIS'21) 2021.02.16 Online
- 渡邉 健太郎, 加登 翔太, 磯部 知弥, 緒方 瑛人, 田坂 佳資, 上野 浩生, 南谷 泰仁, 田中 洋子, 白石 友一, 千葉 健一, 梅田 雄嗣, 樋渡 光輝, 宮野 悟, 小川 誠司, 滝田 順子. 小児・AYA腫瘍の最近の進歩 難治性骨肉腫における糖鎖遺伝子の役割と治療標的としての可能性. 第79回日本癌学会学術総会 2020.10.03 リーガロイヤルホテル広島
- 渡邉 健太郎、加登 翔太、磯部 知弥、緒方 瑛人、田坂 佳資、上 野 浩生、南谷 泰仁、田中 洋子、白石 友一、千葉 健一、梅田 雄嗣、樋渡 光輝、宮野 悟、小川 誠司、滝田 順子. 難治性骨肉腫における糖鎖遺伝子の役割と治療標的としての可能性. 2020.10.03 リーガロイヤルホテル広島
- 古川 洋一、山口 貴世志、笠島 理加、清水 英悟、高根 希世子、 山口 類、井元 清哉、宮野 悟、池上 恒雄. 消化管遺伝性腫瘍の最前線. 第79回日本癌学会学術総会 2020.10.03
- 宮野 悟. AIとがん研究・医療との対話 - がん研究・医療のための自然言語処理と説明可能 AI. 第79回日本癌学会学術総会 2020.10.02 リーガロイヤルホテル広島
- 垣内 伸之、内野 基、木原 多佳子、赤木 宏太朗、井上 善景、横 山 顕礼、平野 智紀、廣田 誠一、池内 浩基、竹内 理、宮野 悟、妹尾 浩、小川 誠司. 潰瘍性大腸炎における大腸上皮クローン進化から明らかとなった大腸 がんの脆弱性. 第79回日本癌学会学術総会 2020.10.02 リーガロイヤルホテル広島
- 井上 善景, 垣内 伸之, 吉田 健一, 塩澤 裕介, 竹内 康英, 千葉 健一, 吉里 哲一, 長山 聡, 宮野 悟, 坂井 義治, 小川 誠司. POLE遺伝子に変異を持つ大腸癌における免疫回避機構. 日本癌学会総会記事 2020.10.01
- 南谷 泰仁, 牧島 秀樹, 竹田 淳恵, 桃沢 幸秀, 佐伯 龍之介, 宮崎 泰司, 石川 隆之, 大屋敷 一馬, Hellstroem Lindberg Eva, Mario Cazzola, Torsten Haferlach, 鎌谷 洋一郎, 久保 充明, 宮野 悟, 小川 誠司. 骨髄系腫瘍におけるDDX41変異の意義. 日本癌学会総会記事 2020.10.01
- 古川 洋一, 山口 貴世志, 笠島 理加, 清水 英悟, 高根 希世子, 山口 類, 井元 清哉, 宮野 悟, 池上 恒雄. 遺伝性腫瘍-最新情報と今後の方向性 消化管遺伝性腫瘍の最前線. 日本癌学会総会記事 2020.10.01
- 垣内 伸之, 横山 顕礼, 内野 基, 木原 多佳子, 赤木 宏太朗, 井上 善景, 平野 智紀, 廣田 誠一, 池内 浩基, 竹内 理, 宮野 悟, 妹尾 浩, 小川 誠司. 遺伝子変異クローンによる食道および大腸組織の再構築の解明. 第79回日本癌学会学術総会 2020.10.01
- 笠島 理加, 鈴木 理樹, 清水 英悟, 玉田 嘉紀, 新井田 厚司, 山口 類, 井元 清哉, 古川 洋一, 宮野 悟, 横瀬 智之, 宮城 洋平. 胎児性肺癌における遺伝子ネットワーク解析. 日本癌学会総会記事 2020.10.01
- 大津 敬, 笠島 理加, 鷲見 公太, 廣島 幸彦, 片山 琴絵, 清水 英悟, 山口 類, 井元 清哉, 宮野 悟, 河村 大輔, 石川 俊平, 横瀬 智之, 比留間 徹, 宮城 洋平. 肉腫患者由来ゼノグラフトのRNA発現解析. 日本癌学会総会記事 2020.10.01
- 竹内 康英, 鈴木 啓道, 吉田 健一, 白石 友一, 垣内 伸之, 塩澤 裕介, 井上 善景, 千葉 健一, 牧島 秀樹, 宮野 悟, 羽賀 博典, Damm Frederik, 小川 誠司. 粘液線維肉腫にみられるTP53の異常と著明な遺伝的不安定性. 日本癌学会総会記事 2020.10.01
- 鎌谷 高志, 平田 真, 植田 幸嗣, 片山 琴絵, 山口 類, 松原 大祐, 藤田 征志, 高澤 豊, 山下 享子, 中村 卓郎, 井元 清哉, 宮野 悟, 中川 英刀, 松田 浩一, 角田 達彦. 粘液型脂肪肉腫におけるがん微小環境の免疫学的解析. 日本癌学会総会記事 2020.10.01
- 平野 智紀, 垣内 伸之, 竹内 康英, 増井 俊彦, 上本 伸二, 南口 早智子, 羽賀 博典, 白石 友一, 宮野 悟, 宇座 徳光, 児玉 裕三, 妹尾 浩, 千葉 勉, 小川 誠司. 異時性多発膵癌の遺伝子解析. 日本癌学会総会記事 2020.10.01
- 越智 陽太郎, 吉田 健一, 佐々木 光, 細谷 紀子, 塩澤 裕介, 南谷 泰仁, 石川 隆之, 白石 友一, 千葉 健一, 田中 洋子, 真田 昌, 牧島 秀樹, 高折 晃史, 宮野 悟, 三谷 絹子, 小川 誠司. 慢性骨髄性白血病急性転化の遺伝学的機序と予後. 日本癌学会総会記事 2020.10.01
- 竹田 淳恵, 吉田 健一, 依田 成玄, 南谷 泰仁, 中川 正宏, 越智 陽太郎, 昆 彩奈, 吉里 哲一, 塩澤 裕介, 白石 友一, 千葉 健一, 田中 洋子, 真田 昌, 宮野 悟, 牧島 秀樹, 小川 誠司. 急性赤白血病におけるJAK阻害剤の検討. 日本癌学会総会記事 2020.10.01
- 土方 康基, 横山 和明, 山口 貴世志, 池上 恒雄, 山口 類, 井元 清哉, 内丸 薫, 宮野 悟, 四柳 宏, 東條 有伸, 古川 洋一. 当院における標準治療不応進行がん患者を対象としたクリニカルシーケンスの検討. 日本癌学会総会記事 2020.10.01
- 垣内 伸之, 内野 基, 木原 多佳子, 赤木 宏太朗, 井上 善景, 横山 顕礼, 平野 智紀, 廣田 誠一, 池内 浩基, 竹内 理, 宮野 悟, 妹尾 浩, 小川 誠司. 大腸がんの診断・治療と発がん研究における新しい知見 潰瘍性大腸炎における大腸上皮クローン進化から明らかとなった大腸がんの脆弱性. 日本癌学会総会記事 2020.10.01
- 西村 友美, 垣内 伸之, 吉田 健一, 竹内 康英, 前田 紘奈, 塩澤 裕介, 平田 勝啓, 片岡 竜貴, 桜井 孝規, 馬場 郷子, 竹内 賢吾, 羽賀 博典, 宮野 悟, 戸井 雅和, 小川 誠司. 乳がん微小環境の特性と全身性応答、新しい治療展開 乳管上皮増殖性病変から乳癌へ至るクローン進化. 日本癌学会総会記事 2020.10.01
- 藤井 陽一, 佐藤 悠佑, 鈴木 啓道, 吉里 哲一, 垣内 伸之, 吉田 健一, 千葉 健一, 白石 友一, 西松 寛明, 岡根谷 利一, 真田 昌, 南谷 泰仁, 牧島 秀樹, 宮野 悟, 久米 春喜, 小川 誠司. 上部尿路上皮癌の分子分類と新規バイオマーカー. 日本癌学会総会記事 2020.10.01
- 関口 昌央, 関 正史, 吉田 健一, 田中 水緒, 白井 了太, 宗崎 良太, 白石 友一, 樋渡 光輝, 加藤 元博, 田口 智章, 田中 祐吉, 宮野 悟, 小川 誠司, 滝田 順子. マルチオミックス解析による肝芽腫の治療標的NQO1、ODC1の同定. 日本癌学会総会記事 2020.10.01
- 小笠原 辰樹, 藤井 陽一, 塩澤 裕介, 牧島 秀樹, 中村 英二郎, 田中 知明, 白石 友一, 宮野 悟, 小川 誠司. チアノーゼ性先天性心疾患に伴う多発パラガングリオーマは異なるHIF2α変異を伴う平行進化を示した. 日本癌学会総会記事 2020.10.01
- 佐伯 龍之介, 吉里 哲一, 白石 友一, 千葉 健一, 田中 洋子, 松田 浩一, 村上 善則, 久保 充明, 宮野 悟, 牧島 秀樹, 小川 誠司. クローン造血における遺伝子変異とコピー数異常の統合解析. 日本癌学会総会記事 2020.10.01
- 藤田 征志, 山口 類, 有廣 光司, 島田 周, 宮野 悟, 山上 裕機, 茶山 一彰, 垣見 和宏, 田中 真二, 井元 清哉, 中川 英刀. がん免疫ゲノム解析 肝臓がんの免疫ゲノム解析. 日本癌学会総会記事 2020.10.01
- 藤田 征志、山口 類、有廣 光司、島田 周、宮野 悟、山上 裕 機、茶山 一彰、垣見 和宏、田中 真二、井元 清哉2、中川 英刀. 肝臓がんの免疫ゲノム解析. 第79回日本癌学会学術総会 2020.10.01 リーガロイヤルホテル広島
- 前田 紘奈、垣内 伸之、平野 智紀、竹内 康英、伊藤 孝司、 小川 絵里、塩川 雅広、宇座 徳光、田中 洋子、南谷 泰仁、牧 島 秀樹、上本 伸二、宮野 悟、小川 誠司. 慢性炎症に伴う胆管上皮におけるクローン拡大. 第79回日本癌学会学術総会 2020.10.01 リーガロイヤルホテル広島
- Satoru Miyano. Cancer Big Data Challenges from Genomes to Networks. The 24th International Conference on Research in Computational Molecular Biology (RECOBM 2020) 2020.06.23 Online
- Nobuyuki Kakiuchi, Kenichi Yoshida, Motoi Uchino, Takako Kihara, Akaki Kotaro, Yoshikage Inoue, Kenji Kawada, Satoshi Nagayama, Akira Yokoyama, Tomonori Hirano, Yasuhide Takeuchi, Hiroyuki Miyoshi, Yoshiharu Sakai, Hironori Haga, Seiichi Hirota, Hiroki Ikeuchi, Osamu Takeuchi, Satoru Miyano, Hiroshi Seno, Seishi Ogawa. Analysis of clonal expansion in epithelium affected by ulcerative colitis reveals frequent mutations affecting IL-17 signaling pathway and novel cancer vulnerability. AACR Annual Meeting 2020 2020.04.27 Online, Philadelphia, USA
- Hirano Tomonori, Nobuyuki Kakiuchi, Yasuhide Takeuchi, Yoshikage Inoue, Tomomi Nishimura, Yoichi Fujii, Akira Yokoyama, Hideki Makishima, Toshihiko Masui, Shinji Uemoto, Sachiko Minamiguchi, Hironori Haga, Kenichi Chiba, Hiroko Tanaka, Yuichi Shiraishi, Satoru Miyano, Norimitsu Uza, Yuzo Kodama, Hiroshi Seno, Seishi Ogawa. Genetic analysis of metachronous pancreatic cancers. AACR Annual Meeting 2020 2020.04.27 Online, Philadelphia, USA
- Yoichi Fujii, Yusuke Sato, Hiromichi Suzuki, Tetsuichi Yoshizato, Kenichi Yoshida, Yuichi Shiraishi, Taketo Kawai, Tohru Nakagawa, Hiroaki Nishimatsu, Toshikazu Nishimatsu, Masashi Sanada, Hideki Makishima, Satoru Miyano, Haruki Kume, Seishi Ogawa. Distinct molecular subtypes and a high diagnostic urinary biomarker of upper urinary tract urothelial carcinoma. AACR Annual Meeting 2020 2020.04.27 Online, Philadelphia, USA
- Yoshikage Inoue, Nobuyuki Kakiuchi, Kenichi Yoshida, Yasuhide Takeuchi, Yusuke Shiozawa, Yuichi Shiraishi, Kenichi Chiba, Tetsuichi Yoshizato, Hiroko Tanaka, Ai Okada, Satoshi Nagayama, Satoru Miyano, Yoshiharu Sakai, Seishi Ogawa. Frequent genomic alterations to evade the immune system in colorectal cancer with POLE gene mutation. AACR Annual Meeting 2020 2020.04.27 Online, Philadelphia, USA
- Yasuhide Takeuchi, Annegret Kunitz, Adriane Halik, Hiromichi Suzuki, Kenichi Yoshida, Yuichi Shiraishi, Kenichi Chiba, Hiroko Tanaka, Nobuyuki Kakiuchi, Yusuke Shiozawa, Akira Yokoyama, Yoshikage Inoue, Tetsuichi Yoshizato, Kosuke Aoki, Yoichi Fujii, Yasuhito Nannya, Hideki Makishima, Satoru Miyano, Hironori Haga, Frederik Damm, Seishi Ogawa. Frequent abnormalities in TP53 and increased genetic instability in myxofibrosarcoma. AACR Annual Meeting 2020 2020.04.27 Online, Philadelphia, USA
- 第16回ヘルシーソサエティ賞(パイオニア部門),ヘルシー・ソサエティ賞事務局,2020年11月
Major Past Research Achievements
※Bold belong to this field
- ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium. Pan-cancer analysis of whole genomes. Nature. 578(7793):82-93, 2020.
- Kakiuchi N, Yoshida K, Uchino M, Kihara T, Akaki K, Inoue Y, Kawada K, Nagayama S, Yokoyama A, Yamamoto S, Matsuura M, Horimatsu T, Hirano T, Goto N, Takeuchi Y, Ochi Y, Shiozawa Y, Kogure Y, Watatani Y, Fujii Y, Kim SK, Kon A, Kataoka K, Yoshizato T, MM Nakagawa, Yoda A, Nannya Y, Makishima H, Shiraishi Y, Chiba K, Tanaka H, Sanada M, Sugihara E, Sato T, Maruyama T, Miyoshi H, Taketo MM, Oishi J, Inagaki R, Ueda Y, Okamoto S, Okajima H, Sakai Y, Sakurai T, Haga H, Hirota S, Ikeuchi H, Nakase H, Marusawa H, Chiba T, Takeuchi O, Miyano S, Seno H, Ogawa S. Frequent mutations converging into NFKBIZ signalling in ulcerative colitis. Nature. 577:260–265, 2020.
- Ochi Y, Kon A, Sakata T, Nakagawa MM, Nakazawa N, Kakuta M, Kataoka K, Koseki H, Nakayama M, Morishita D, Tsuruyama T, Saiki R, Yoda A, Okuda R, Yoshizato T, Yoshida K, Shiozawa Y, Nannya Y, Kotani S, Kogure Y, Kakiuchi N, Nishimura T, Makishima H, Malcovati L, Yokoyama A, Takeuchi K, Sugihara E, Sato TA, Sanada M, Takaori-Kondo A, Cazzola M, Kengaku M, Miyano S, Shirahige K, Suzuki HI, Ogawa S. Combined cohesin-Runx1 deficiency synergistically perturbs chromatin looping and causes myelodysplastic syndromes. Cancer Discov. pii: CD-19-0982. doi: 10.1158/2159-8290, 2020.
- Yokoyama A, Kakiuchi N, Yoshizato T, Nannya Y, Suzuki H, Takeuchi Y, Shiozawa Y, Sato Y, Aoki K, Kim SK, Fujii Y, Yoshida K, Kataoka K, Nakagawa MM, Inoue Y, Hirano T, Shiraishi Y, Chiba K, Tanaka H, Sanada M, Nishikawa Y, Amanuma Y, Ohashi S, Aoyama I, Horimatsu T, Miyamoto S, Tsunoda S, Sakai Y, Narahara M, Brown JB, Sato Y, Sawada G, Mimori K, Minamiguchi S, Haga H, Seno H, Miyano S, Makishima H, Muto M, Ogawa S. Age-related remodelling of oesophageal epithelia by mutated cancer drivers. Nature. 565(7739):312-317, 2019.
- Okuno Y, Murata T, Sato Y, Muramatsu H, Ito Y, Watanabe T, Okuno T, Murakami N, Yoshida K, Sawada A, Inoue M, Kawa K, Seto M, Ohshima K, Shiraishi Y, Chiba K, Tanaka H, Miyano S, Narita Y, Yoshida M, Goshima F, Kawada JI, Nishida T, Kiyoi H, Kato S, Nakamura S, Morishima S, Yoshikawa T, Fujiwara S, Shimizu N, Isobe Y, Noguchi M, Kikuta A, Iwatsuki K, Takahashi Y, Kojima S, Ogawa S, Kimura H. Defective Epstein-Barr virus in chronic active infection and haematological malignancy. Nature Microbiology. 4(3):404-413, 2019; Publisher Correction: Nature Microbiology. 2019 Mar;4(3):544.
- Matsuno Y, Atsumi Y, Shimizu A, Katayama K, Fujimori H, Hyodo M, Minakawa Y, Nakatsu Y, Kaneko S, Hamamoto R, Shimamura T, Miyano S, Tsuzuki T, Hanaoka F, Yoshioka KI. Replication stress triggers microsatellite destabilization and hypermutation leading to clonal expansion in vitro. Nature Commun. 10(1):3925, 2019.
- Kajino T, Shimamura T, Gong S, Yanagisawa K, Ida L, Nakatochi M, Griesing S, Shimada Y, Kano K, Suzuki M, Miyano S, Takahashi T. Divergent lncRNA MYMLR regulates MYC by eliciting DNA looping and promoter-enhancer interaction. EMBO J. 8(17):e98441, 2019.
- Saito T, Niida A, Uchi R, Hirata H, Komatsu H, Sakimura S, Hayashi S, Nambara S, Kuroda Y, Ito S, Eguchi H, Masuda T, Sugimachi K, Tobo T, Nishida H, Daa T, Chiba K, Shiraishi Y, Yoshizato T, Kodama M, Okimoto T, Mizukami K, Ogawa R, Okamoto K, Shuto M, Fukuda K, Matsui Y, Shimamura T, Hasegawa T, Doki Y, Nagayama S, Yamada K, Kato M, Shibata T, Mori M, Aburatani H, Murakami K, Suzuki Y, Ogawa S, Miyano S, Mimori K. A temporal shift of the evolutionary principle shaping intratumor heterogeneity in colorectal cancer. Nature Commun. 9(1):2884, 2018.
- Shiozawa Y, Malcovati L, Gallì A, Sato-Otsubo A, Kataoka K, Sato Y, Watatani Y, Suzuki H, Yoshizato T, Yoshida K, Sanada M, Makishima H, Shiraishi Y, Chiba K, Hellström-Lindberg E, Miyano S, Ogawa S, Cazzola M. Aberrant splicing and defective mRNA production induced by somatic spliceosome mutations in myelodysplasia. Nature Commun. 9(1):3649, 2018.
- da Silva-Coelho P, Kroeze LI, Yoshida K, Koorenhof-Scheele TN, Knops R, van de Locht LT, de Graaf AO, Massop M, Sandmann S, Dugas M, Stevens-Kroef MJ, Cermak J, Shiraishi Y, Chiba K, Tanaka H, Miyano S, de Witte T, Blijlevens NMA, Muus P, Huls G, van der Reijden BA, Ogawa S, Jansen JH. Clonal evolution in myelodysplastic syndromes. Nature Commun. 8:15099, 2017.
- Makishima H, Yoshizato T, Yoshida K, Sekeres MA, Radivoyevitch T, Suzuki H, Przychodzen B, Nagata Y, Meggendorfer M, Sanada M, Okuno Y, Hirsch C, Kuzmanovic T, Sato Y, Sato-Otsubo A, LaFramboise T, Hosono N, Shiraishi Y, Chiba K, Haferlach C, Kern W, Tanaka H, Shiozawa Y, Gómez-Seguí I, Husseinzadeh HD, Thota S, Guinta KM, Dienes B, Nakamaki T, Miyawaki S, Saunthararajah Y, Chiba S, Miyano S, Shih LY, Haferlach T, Ogawa S, Maciejewski JP. Dynamics of clonal evolution in myelodysplastic syndromes. Nature Genetics. 49(2):204-212, 2017.
- Seki M, Kimura S, Isobe T, Yoshida K, Ueno H, Nakajima-Takagi Y, Wang C, Lin L, Kon A, Suzuki H, Shiozawa Y, Kataoka K, Fujii Y, Shiraishi Y, Chiba K, Tanaka H, Shimamura T, Masuda K, Kawamoto H, Ohki K, Kato M, Arakawa Y, Koh K, Hanada R, Moritake H, Akiyama M, Kobayashi R, Deguchi T, Hashii Y, Imamura T, Sato A, Kiyokawa N, Oka A, Hayashi Y, Takagi M, Manabe A, Ohara A, Horibe K, Sanada M, Iwama A, Mano H, Miyano S, Ogawa S, Takita J. Recurrent SPI1 (PU.1) fusions in high-risk pediatric T cell acute lymphoblastic leukemia. Nature Genetics. 49(8):1274-1281, 2017.
- Fujimoto A, Furuta M, Totoki Y, Tsunoda T, Kato M, Shiraishi Y, Tanaka H, Taniguchi H, Kawakami Y, Ueno M, Gotoh K, Ariizumi S, Wardell CP, Hayami S, Nakamura T, Aikata H, Arihiro K, Boroevich KA, Abe T, Nakano K, Maejima K, Sasaki-Oku A, Ohsawa A, Shibuya T, Nakamura H, Hama N, Hosoda F, Arai Y, Ohashi S, Urushidate T, Nagae G, Yamamoto S, Ueda H, Tatsuno K, Ojima H, Hiraoka N, Okusaka T, Kubo M, Marubashi S, Yamada T, Hirano S, Yamamoto M, Ohdan H, Shimada K, Ishikawa O, Yamaue H, Chayama K, Miyano S, Aburatani H, Shibata T, Nakagawa H. Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nature Genetics. 48(5):500-509, 2016. Erratum: Nature Genetics. 48(6):700. doi: 10.1038/ng0616-700a.
- Kataoka K, Shiraishi Y, Takeda Y, Sakata S, Matsumoto M, Nagano S, Maeda T, Nagata Y, Kitanaka A, Mizuno S, Tanaka H, Chiba K, Ito S, Watatani Y, Kakiuchi N, Suzuki H, Yoshizato T, Yoshida K, Sanada M, Itonaga H, Imaizumi Y, Totoki Y, Munakata W, Nakamura H, Hama N, Shide K, Kubuki Y, Hidaka T, Kameda T, Masuda K, Minato N, Kashiwase K, Izutsu K, Takaori-Kondo A, Miyazaki Y, Takahashi S, Shibata T, Kawamoto H, Akatsuka Y, Shimoda K, Takeuchi K, Seya T, Miyano S, Ogawa S. Aberrant PD-L1 expression through 3'-UTR disruption in multiple cancers. Nature. 534(7607):402-406, 2016.
- Fujimoto A, Furuta M, Shiraishi Y, Gotoh K, Kawakami Y, Arihiro K, Nakamura T, Ueno M, Ariizumi SI, Hai Nguyen H, Shigemizu D, Abe T, Boroevich KA, Nakano K, Sasaki A, Kitada R, Maejima K, Yamamoto Y, Tanaka H, Shibuya T, Shibata T, Ojima H, Shimada K, Hayami S, Shigekawa Y, Aikata H, Ohdan H, Marubashi S, Yamada T, Kubo M, Hirano S, Ishikawa O, Yamamoto M, Yamaue H, Chayama K, Miyano S, Tsunoda T, Nakagawa H. Whole-genome mutational landscape of liver cancers displaying biliary phenotype reveals hepatitis impact and molecular diversity. Nature Commun. 6:6120, 2015.
- Kataoka K, Nagata Y, Kitanaka A, Shiraishi Y, Shimamura T, Yasunaga J, Totoki Y, Chiba K, Sato-Otsubo A, Nagae G, Ishii R, Muto S, Kotani S, Watatani Y, Takeda J, Sanada M, Tanaka H, Suzuki H, Sato Y, Shiozawa Y, Yoshizato T, Yoshida K, Makishima H, Iwanaga M, Ma G, Nosaka K, Hishizawa M, Itonaga H, Imaizumi Y, Munakata W, Ogasawara H, Sato T, Sasai K, Muramoto K, Penova M, Kawaguchi T, Nakamura H, Hama N, Shide K, Kubuki Y, Hidaka T, Kameda T, Nakamaki T, Ishiyama K, Miyawaki S, Yoon SS, Tobinai K, Miyazaki Y, Takaori-Kondo A, Matsuda F, Takeuchi K, Nureki O, Aburatani H, Watanabe T, Shibata T, Matsuoka M, Miyano S, Shimoda K, Ogawa S. Integrated molecular analysis of adult T cell leukemia/lymphoma. Nature Genetics. 47(11):1304-1315, 2015.
- Madan V, Kanojia D, Li J, Okamoto R, Sato-Otsubo A, Kohlmann A, Sanada M, Grossmann V, Sundaresan J, Shiraishi Y, Miyano S, Thol F, Ganser A, Yang H, Haferlach T, Ogawa S, Koeffler HP. Aberrant splicing of U12-type introns is the hallmark of ZRSR2 mutant myelodysplastic syndrome. Nature Commun. 6:6042, 2015.
- Polprasert C, Schulze I, Sekeres MA, Makishima H, Przychodzen B, Hosono N, Singh J, Padgett RA, Gu X, Phillips JG, Clemente M, Parker Y, Lindner D, Dienes B, Jankowsky E, Saunthararajah Y, Du Y, Oakley K, Nguyen N, Mukherjee S, Pabst C, Godley LA, Churpek JE, Pollyea DA, Krug U, Berdel WE, Klein HU, Dugas M, Shiraishi Y, Chiba K, Tanaka H, Miyano S, Yoshida K, Ogawa S, Müller-Tidow C, Maciejewski JP. Inherited and somatic defects in DDX41 in myeloid neoplasms. Cancer Cell. 27(5):658-670, 2015.
- Seki M, Nishimura R, Yoshida K, Shimamura T, Shiraishi Y, Sato Y, Kato M, Chiba K, Tanaka H, Hoshino N, Nagae G, Shiozawa Y, Okuno Y, Hosoi H, Tanaka Y, Okita H, Miyachi M, Souzaki R, Taguchi T, Koh K, Hanada R, Kato K, Nomura Y, Akiyama M, Oka A, Igarashi T, Miyano S, Aburatani H, Hayashi Y, Ogawa S, Takita J. Integrated genetic and epigenetic analysis defines novel molecular subgroups in rhabdomyosarcoma. Nature Commun. 6:7557, 2015.
- Suzuki H, Aoki K, Chiba K, Sato Y, Shiozawa Y, Shiraishi Y, Shimamura T, Niida A, Motomura K, Ohka F, Yamamoto T, Tanahashi K, Ranjit M, Wakabayashi T, Yoshizato T, Kataoka K, Yoshida K, Nagata Y, Sato-Otsubo A, Tanaka H, Sanada M, Kondo Y, Nakamura H, Mizoguchi M, Abe T, Muragaki Y, Watanabe R, Ito I, Miyano S, Natsume A, Ogawa S. Mutational landscape and clonal architecture in grade II and III gliomas. Nature Genetics. 47(5):458-468, 2015.
- Yoshizato T, Dumitriu B, Hosokawa K, Makishima H, Yoshida K, Townsley D, Sato-Otsubo A, Sato Y, Liu D, Suzuki H, Wu CO, Shiraishi Y, Clemente MJ, Kataoka K, Shiozawa Y, Okuno Y, Chiba K, Tanaka H, Nagata Y, Katagiri T, Kon A, Sanada M, Scheinberg P, Miyano S, Maciejewski JP, Nakao S, Young NS, Ogawa S. Somatic mutations and clonal hematopoiesis in aplastic anemia. N Engl J Med. 373(1):35-47, 2015.
- Arima C, Kajino T, Tamada Y, Imoto S, Shimada Y, Nakatochi M, Suzuki M, Isomura H, Yatabe Y, Yamaguchi T, Yanagisawa K, Miyano S, Takahashi T. Lung adenocarcinoma subtypes definable by lung development-related miRNA expression profiles in association with clinicopathologic features. Carcinogenesis. 35(10): 2224-2231, 2014.
- Sakata-Yanagimoto M, Enami T, Yoshida K, Shiraishi Y, Ishii R, Miyake Y, Muto H, Tsuyama N, Sato-Otsubo A, Okuno Y, Sakata S, Kamada Y, Nakamoto-Matsubara R, Tran NB, Izutsu K, Sato Y, Ohta Y, Furuta J, Shimizu S, Komeno T, Sato Y, Ito T, Noguchi M, Noguchi E, Sanada M, Chiba K, Tanaka H, Suzukawa K, Nanmoku T, Hasegawa Y, Nureki O, Miyano S, Nakamura N, Takeuchi K, Ogawa S, Chiba S. Somatic RHOA mutation in angioimmunoblastic T cell lymphoma. Nature Genetics. 46(2):171-175, 2014.
- Sato Y, Maekawa S, Ishii R, Sanada M, Morikawa T, Shiraishi Y, Yoshida K, Nagata Y, Sato-Otsubo A, Yoshizato T, Suzuki H, Shiozawa Y, Kataoka K, Kon A, Aoki K, Chiba K, Tanaka H, Kume H, Miyano S, Fukayama M, Nureki O, Homma Y, Ogawa S. Recurrent somatic mutations underlie corticotropin-independent Cushing's syndrome. Science. 344(6186):917-920, 2014.
- Kon A, Shih LY, Minamino M, Sanada M, Shiraishi Y, Nagata Y, Yoshida K, Okuno Y, Bando M, Nakato R, Ishikawa S, Sato-Otsubo A, Nagae G, Nishimoto A, Haferlach C, Nowak D, Sato Y, Alpermann T, Nagasaki M, Shimamura T, Tanaka H, Chiba K, Yamamoto R, Yamaguchi T, Otsu M, Obara N, Sakata-Yanagimoto M, Nakamaki T, Ishiyama K, Nolte F, Hofmann WK, Miyawaki S, Chiba S, Mori H, Nakauchi H, Koeffler HP, Aburatani H, Haferlach T, Shirahige K, Miyano S, Ogawa S. Recurrent mutations in multiple components of the cohesin complex in myeloid neoplasms. Nature Genetics. 45(10):1232-1237, 2013.
- Makishima H, Yoshida K, Nguyen N, Przychodzen B, Sanada M, Okuno Y, Ng KP, Gudmundsson KO, Vishwakarma BA, Jerez A, Gomez-Segui I, Takahashi M, Shiraishi Y, Nagata Y, Guinta K, Mori H, Sekeres MA, Chiba K, Tanaka H, Muramatsu H, Sakaguchi H, Paquette RL, McDevitt MA, Kojima S, Saunthararajah Y, Miyano S, Shih LY, Du Y, Ogawa S, Maciejewski JP. Somatic SETBP1 mutations in myeloid malignancies. Nature Genetics. 45(8):942-946, 2013.
- Sakaguchi H, Okuno Y, Muramatsu H, Yoshida K, Shiraishi Y, Takahashi M, Kon A, Sanada M, Chiba K, Tanaka H, Makishima H, Wang X, Xu Y, Doisaki S, Hama A, Nakanishi K, Takahashi Y, Yoshida N, Maciejewski JP, Miyano S, Ogawa S, Kojima S. Exome sequencing identifies secondary mutations of SETBP1 and JAK3 in juvenile myelomonocytic leukemia. Nature Genetics. 45(8):937-941, 2013.
- Sato Y, Yoshizato T, Shiraishi Y, Maekawa S, Okuno Y, Kamura T, Shimamura T, Sato-Otsubo A, Nagae G, Suzuki H, Nagata Y, Yoshida K, Kon A, Suzuki Y, Chiba K, Tanaka H, Niida A, Fujimoto A, Tsunoda T, Morikawa T, Maeda D, Kume H, Sugano S, Fukayama M, Aburatani H, Sanada M, Miyano S, Homma Y, Ogawa S. Integrated molecular analysis of clear-cell renal cell carcinoma. Nature Genetics. 45(8):860-867, 2013.
- Yoshida K, Toki T, Okuno Y, Kanezaki R, Shiraishi Y, Sato-Otsubo A, Sanada M, Park MJ, Terui K, Suzuki H, Kon A, Nagata Y, Sato Y, Wang R, Shiba N, Chiba K, Tanaka H, Hama A, Muramatsu H, Hasegawa D, Nakamura K, Kanegane H, Tsukamoto K, Adachi S, Kawakami K, Kato K, Nishimura R, Izraeli S, Hayashi Y, Miyano S, Kojima S, Ito E, Ogawa S. The landscape of somatic mutations in Down syndrome-related myeloid disorders. Nature Genetics. 45:1293–1299, 2013. Erratum in: Nature Genetics. 45(12):1516, 2013.
- Yoshimaru T, Komatsu M, Matsuo T, Chen YA, Murakami Y, Mizuguchi K, Mizohata E, Inoue T, Akiyama M, Yamaguchi R, Imoto S, Miyano S, Miyoshi Y, Sasa M, Nakamura Y, Katagiri T. Targeting BIG3-PHB2 interaction to overcome tamoxifen resistance in breast cancer cells. Nature Commun. 4:2443, 2013.
- Fujimoto A, Totoki Y, Abe T, Boroevich KA, Hosoda F, Hai Nguyen H, Aoki M, Hosono N , Kubo M, Miya F, Arai Y, Takahashi H, Shirakihara T, Nagasaki M, Shibuya T, Nakano K, Watanabe-Makino K, Tanaka H, Nakamura H, Kusuda J, Ojima H , Shimada K, Okusaka T, Ueno M, Shigekawa Y, Kawakami Y, Arihiro K, Ohdan H, Gotoh K, Ishikawa O, Ariizumi S, Yamamoto M, Yamada T, Chayama K, Kosuge T, Yamaue H, Kamatani N, Miyano S, Nakagama H, Nakamura Y, Tsunoda T, Shibata T, Nakagawa H. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nature Genetics . 44(7):760-764, 2012 .
- Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, Sato Y, Sato-Otsubo A, Kon A, Nagasaki M, Chalkidis G, Suzuki Y, Shiosaka M, Kawahata R, Yamaguchi T, Otsu M, Obara N, Sakata-Yanagimoto M, Ishiyama K, Mori H, Nolte F, Hofmann WK, Miyawaki S, Sugano S, Haferlach C, Koeffler HP, Shih LY, Haferlach T, Chiba S, Nakauchi H, Miyano S, Ogawa S. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 478(7367): 64-69, 2011.
- Fujimoto A, Nakagawa H, Hosono N, Nakano K, Abe G, Boroevich KA, Nagasaki M, Yamaguchi R, Shibuya T, Kubo M, Miyano S, Nakamura Y, Tsunoda T. Whole-genome sequencing and comprehensive variant analysis of a Japanese individual using massively parallel sequencing. Nature Genetics. 42: 931–936, 2010.
- Fujita M, Yamaguchi R, Hasegawa T, Shimada S, Arihiro K, Hayashi S, Maejima K, Nakano K, Fujimoto A, Ono A, Aikata H, Ueno M, Hayami S, Tanaka H, Miyano S, Yamaue H, Chayama K, Kakimi K, Tanaka S, Imoto S, Nakagawa H. Classification of primary liver cancer with immunosuppression mechanisms and correlation with genomic alterations. EBioMedicine. 2020 Mar;53:102659. doi: 10.1016/j.ebiom.2020.102659.
- Fujimoto A, Fujita M, Hasegawa T, Wong JH, Maejima K, Oku-Sasaki A, Nakano K, Shiraishi Y, Miyano S, Yamamoto G, Akagi K, Imoto S, Nakagawa H. Comprehensive analysis of indels in whole-genome microsatellite regions and microsatellite instability across 21 cancer types. Genome Res. 2020 Mar 24. doi: 10.1101/gr.255026.119.
- Hasegawa T, Yamaguchi R, Kakuta M, Sawada K, Kawatani K, Murashita K, Nakaji S, Imoto S. Prediction of blood test values under different lifestyle scenarios using time-series electronic health record. PLoS One. 15(3):e0230172, 2020.
- Sato N, Kakuta M, Uchino E, Hasegawa T, Kojima R, Kobayashi W, Sawada K, Tamura Y, Tokuda I, Imoto S, Nakaji S, Murashita K, Yanagita M, Okuno Y. The relationship between cigarette smoking and the tongue microbiome in an East Asian population. J Oral Microbiol. 12(1):1742527, 2020.
- Sato N, Kakuta M, Hasegawa T, Yamaguchi R, Uchino E, Kobayashi W, Sawada K, Tamura Y, Tokuda I, Murashita K, Nakaji S, Imoto S, Yanagita M, Okuno Y. Metagenomic analysis of bacterial species in tongue microbiome of current and never smokers. NPJ Biofilms Microbiomes. 6(1):11, 2020.
- Misawa K, Hasegawa T, Mishima E, Jutabha P, Ouchi M, Kojima K, Kawai Y, Matsuo M, Anzai N, Nagasaki M. Contribution of rare variants of the SLC22A12 gene to the missing heritability of serum urate levels. Genetics. 214(4):1079-1090, 2020.
- Ito S, Yadome M, Nishiki T, Ishiduka S, Yamaguchi R, Miyano S. Virtual Grid Engine: A simulated grid engine environment for large-scale supercomputers. BMC Bioinformatics. 20(Suppl 16):591, 2019.
- Moriyama T, Imoto S, Hayashi S, Shiraishi Y, Miyano S, Yamaguchi R. A Bayesian model integration for mutation calling through data partitioning. Bioinformatics. 35(21):4247-4254, 2019.
- Hayashi S, Moriyama T, Yamaguchi R, Mizuno S, Komura M, Miyano S, Nakagawa H, Imoto S. ALPHLARD-NT: Bayesian method for human leukocyte antigen genotyping and mutation calling through simultaneous analysis of normal and tumor whole-genome sequence data. J Comput Biol. 26(9):923-937, 2019.
- Hayashi S, Yamaguchi R, Mizuno S, Komura M, Miyano S, Nakagawa H, Imoto S. ALPHLARD: a Bayesian method for analyzing HLA genes from whole genome sequence data. BMC Genomics. 19(1):790, 2018.
- Shiraishi Y, Kataoka K, Chiba K, Okada A, Kogure Y, Tanaka H, Ogawa S, Miyano S. A comprehensive characterization of cis-acting splicing-associated variants in human cancer. Genome Res. 28(8):1111-1125, 2018.
- Moriyama T, Shiraishi Y, Chiba K, Yamaguchi R, Imoto S, Miyano S. OVarCall: Bayesian mutation calling method utilizing overlapping paired-end reads. IEEE Trans Nanobioscience. 16(2): 116-122, 2017.
- Zhang YZ, Yamaguchi R, Imoto S, Miyano S. Sequence-specific bias correction for RNA-seq data using recurrent neural networks. BMC Genomics. 18(Suppl 1):1044, 2017.
- Zhang YZ, Imoto S, Miyano S, Yamaguchi R. Reconstruction of high read-depth signals from low-depth whole genome sequencing data using deep learning. BIBM 2017: 1227-1232, 2017.
- Hasegawa T, Kojima K, Kawai Y, Misawa K, Mimori T, Nagasaki M. AP-SKAT: highly-efficient genome-wide rare variant association test. BMC Genomics. 17(1):745, 2016.
- Kojima K, Kawai Y, Nariai N, Mimori T, Hasegawa T, Nagasaki M. Short tandem repeat number estimation from paired-end reads for multiple individuals by considering coalescent tree. BMC Genomics. 17 Suppl 5:494, 2016.
- Koshiba S, Motoike I, Kojima K, Hasegawa T, Shirota M, Saito T, Saigusa D, Danjoh I, Katsuoka F, Ogishima S, Kawai Y, Yamaguchi-Kabata Y, Sakurai M, Hirano S, Nakata J, Motohashi H, Hozawa A, Kuriyama S, Minegishi N, Nagasaki M, Takai-Igarashi T, Fuse N, Kiyomoto H, Sugawara J, Suzuki Y, Kure S, Yaegashi N, Tanabe O, Kinoshita K, Yasuda J, Yamamoto M. The structural origin of metabolic quantitative diversity. Sci Rep. 6:31463, 2016.
- Chiba K, Shiraishi Y, Nagata Y, Yoshida K, Imoto S, Ogawa S, Miyano S. Genomon ITDetector: A tool for somatic internal tandem duplication detection from cancer genome sequencing data. Bioinformatics. 31(1), 116-118, 2015.
- Ito S, Shiraishi Y, Shimamura T, Chiba K, Miyano S. High performance computing of a fusion gene detection pipeline on the K computer. BIBM 2015: 1441-1447, 2015.
- Shiraishi Y, Sato Y, Chiba K, Okuno Y, Nagata Y, Yoshida K, Shiba N, Hayashi Y, Kume H, Homma Y, Sanada M, Ogawa S, Miyano S. An empirical Bayesian framework for somatic mutation detection from cancer. Nucleic Acids Res. 41(7): e89, 2013.
- Niida A, Imoto S, Shimamura T, Miyano S. Statistical model-based testing to evaluate the recurrence of genomic aberrations. Bioinformatics. 28(12):i115-i120, 2012.
- Shiraishi U, Okada-Hatakeyama M, Miyano S. A rank-based statistical test for measuring synergistic effects between two gene sets. Bioinformatics. 27 (17): 2399-2405, 2011.
- Hasegawa T, Yamaguchi R, Niida A, Miyano S, Imoto S. Ensemble smoothers for inference of hidden states and parameters in combinatorial regulatory model. J Franklin Institute. (5):2916-2933, 2020.
- Niida A, Hasegawa T, Innan H, Shibata T, Mimori K, Miyano S. A unified simulation model for understanding the diversity of cancer evolution. PeerJ. 8:e8842, 2020.
- Niida A, Hasegawa T, Miyano S. Sensitivity analysis of agent-based simulation utilizing massively parallel computation and interactive data visualization. PLoS One. 14(3):e0210678, 2019.
- Park H, Yamada M, Imoto S, Miyano S. Robust sample-specific stability selection with effective error control. J Comput Biol. 26(3):202-217, 2019.
- Park H, Shimamura T, Imoto S, Miyano S. Adaptive NetworkProfiler for identifying cancer characteristic-specific gene regulatory networks. J Comput Biol. 25(2):130-145, 2018.
- Mimori K, Saito T, Niida A, Miyano S. Cancer evolution and heterogeneity. Ann Gastroenterol Surg. 2(5):332-338, 2018.
- Hasegawa T, Kojima K, Kawai Y, Nagasaki M. Time-series filtering for replicated observations via a kernel approximate Bayesian computation. IEEE Transactions on Signal Processing. 66(23):6148-6161, 2018.
- Niida A, Nagayama S, Miyano S, Mimori K. Understanding intratumor heterogeneity by combining genome analysis and mathematical modeling. Cancer Sci. 109(4):884-892, 2018.
- Park H, Niida A, Imoto S, Miyano S. Interaction-based feature selection for uncovering cancer driver genes through copy number-driven expression level. J Comput Biol. 24(2):138-152, 2017.
- Park H, Shiraishi Y, Imoto S, Miyano S. A novel adaptive penalized logistic regression for uncovering biomarker associated with anti-cancer drug sensitivity. IEEE/ACM Trans Comput Biol Bioinform. 14(4):771-782, 2017.
- Matsui Y, Niida A, Uchi R, Mimori K, Miyano S, Shimamura T. phyC: Clustering cancer evolutionary trees. PLoS Comput Biol. 13(5):e1005509, 2017.
- Morita T, Rahman A, Hasegawa T, Ozaki A, Tanimoto T. The potential possibility of symptom checker. Int J Health Policy Manag. 6(10):615-616, 2017.
- Kurashige J, Hasegawa T, Niida A, Sugimachi K, Deng N, Mima K, Uchi R, Sawada G, Takahashi Y, Eguchi H, Inomata M, Kitano S, Fukagawa T, Sasako M, Sasaki H, Sasaki S, Mori M, Yanagihara K, Baba H, Miyano S, Tan P, Mimori K. Integrated molecular profiling of human gastric cancer identifies DDR2 as a potential regulator of peritoneal dissemination. Sci Rep. 6:22371, 2016.
- Hasegawa T, Mori T, Yamaguchi R, Shimamura T, Miyano S, Imoto S, Akutsu T. Genomic data assimilation using a higher moment filtering technique for restoration of gene regulatory networks. BMC Syst Biol. 9:14, 2015.
- Park H, Niida, A, Miyano S, Imoto S. Sparse overlapping group lasso for integrative multi-omics analysis. J Comput Biol. 22(2): 73-84, 2015.
- Park H, Imoto S, Miyano S. Recursive Random Lasso (RRLasso) for identifying anti-cancer drug targets. PLoS One. 10(11):e0141869, 2015.
- Nakata A, Yoshida R, Yamaguchi R, Yamauchi M, Tamada Y, Fujita A, Shimamura T, Imoto S, Higuchi T, Nomura M, Kimura T, Nokihara H, Higashiyama M, Kondoh K, Nishihara H, Tojo A, Yano S, Miyano S, Gotoh N. Elevated β-catenin pathway as a novel target for patients with resistance to EGF receptor targeting drugs. Sci Rep. 5:13076, 2015.
- Hasegawa T, Yamaguchi R, Nagasaki M, Miyano S, Imoto S. Inference of gene regulatory networks incorporating multi-source biological knowledge via a state space model with L1 regularization. PLoS One. 9(8):e105942, 2014.
- Hasegawa T, Nagasaki M, Yamaguchi R, Imoto S, Miyano S. An efficient method of exploring simulation models by assimilating literature and biological observational data. Biosystems. 121:54-66, 2014.
- Hasegawa T, Mori T, Yamaguchi R, Imoto S, Miyano S, Akutsu T. An efficient data assimilation schema for restoration and extension of gene regulatory networks using time-course observation data. J Comput Biol. 21(11):785-798, 2014.
- Park H, Shimamura T, Miyano S, Imoto S. Robust prediction of anti-cancer drug sensitivity and sensitivity-specific biomarker. PLoS One. 9(1): e108990, 2014.
- Yamauchi M, Yamaguchi R, Nakata A, Kohno T, Nagasaki M, Shimamura T, Imoto S, Saito A, Ueno K, Hatanaka Y, Yoshida R, Higuchi T, Nomura M, Beer DG, Yokota J, Miyano S, Gotoh N. Epidermal growth factor receptor tyrosine kinase defines critical prognostic genes of stage I lung adenocarcinoma. PLoS One. 7(9): e43923, 2012.
- Shimamura T, Imoto S, Shimada Y, Hosono Y, Niida A, Nagasaki M, Yamaguchi R, Takahashi T, Miyano S. A novel network profiling analysis reveals system changes in epithelial-mesenchymal transition. PLoS One. 6(6): e20804, 2011.
- Tamada Y, Imoto S, Miyano S. Parallel algorithm for learning optimal Bayesian network structure. J Machine Learning Research. 12: 2437-2459, 2011.
- Tamada Y, Imoto S, Araki H, Nagasaki M, Print C, Charnock-Jones DS, Miyano S. Estimating genome-wide gene networks using nonparametric Bayesian network models on massively parallel computers. IEEE/ACM Transactions on Computational Biology and Bioinformatics. 8(3): 683 - 697, 2011. Tamada Y, Shimamura T, Yamaguchi R, Imoto S, Nagasaki M, Miyano S. SiGN: large-scale gene network estimation environment for high performance computing. Genome Informatics. 25: 40-52, 2011.
- Tamada Y, Yamaguchi R, Imoto S, Hirose O, Yoshida R, Nagasaki M, Miyano S. SiGN-SSM: open source parallel software for estimating gene networks with state space models. Bioinformatics. 27: 1172-1173, 2011.
- Kojima K, Perrier E, Imoto S, Miyano S. Optimal search on clustered structural constraint for learning Bayesian network structure. J Machine Learning Research. 11: 285−310, 2010.
- Yamaguchi R, Imoto S, Yamauchi M, Nagasaki M, Yoshida R, Shimamura T, Hatanaka Y, Ueno K, Higuchi T, Gotoh N, Miyano S. Predicting differences in gene regulatory systems by state space models. Genome Informatics. 21:101-113, 2008.
- Perrier E, Imoto S, Miyano S. Finding optimal Bayesian network given a super-structure. J Machine Learning Research. 9: 2251-2286, 2008.
- Yamaguchi R, Yoshida R, Imoto S, Higuchi T, Miyano S. Finding module-based gene networks with state-space models? Mining high-dimensional and short time-course gene expression data. IEEE Signal Processing Magazine. 24(1): 37-46, 2007.
- Imoto S, Tamada Y, Araki H, Yasuda K, Print CG, Charnock-Jones SD, Sanders D, Savoie CJ, Tashiro K, Kuhara S, Miyano S. Computational strategy for discovering druggable gene networks from genome-wide RNA expression profiles. Pacific Symposium on Biocomputing. 11: 559-571, 2006.
- Ott S, Hansen A, Kim S-Y, Miyano S. Superiority of network motifs over optimal networks and an application to the revelation of gene network evolution. Bioinformatics. 21(2): 227-238, 2005.
- Imoto S, Goto T, Miyano S. Estimation of genetic networks and functional structures between genes by using Bayesian network and nonparametric regression. Pacific Symposium on Biocomputing. 7:175-186, 2002.
- Anderson WP; Global Life Science Data Resources Working Group: Apweiler R, Bateman A, Bauer GA, Berman H, Blake JA, Blomberg N, Burley SK, Cochrane G, Di Francesco V, Donohue T, Durinx C, Game A, Green E, Gojobori T, Goodhand P, Hamosh A, Hermjakob H, Kanehisa M, Kiley R, McEntyre J, McKibbin R, Miyano S, Pauly B, Perrimon N, Ragan MA, Richards G, Teo YY, Westerfield M, Westhof E, Lasko PF. Data management: A global coalition to sustain core data. Nature. 543(7644):179, 2017.

Department of AI Technology Development

Department of Biostatistics

Department of Data Science Algorithm Design and Analysis


