Department of Data Science Algorithm Design and Analysis
Our research focuses on developing algorithms and data structures for efficiently processing and analyzing large scale data.
Members

Hideo Bannai
Professor
Diptarama Hendrian
Research
String Algorithmics
Algorithms are an essential component of data science, especially for handling large data sets. Our aim is to design algorithms and data structures that are both effective and efficient, to help manage and analyze various types of large-scale medical data such as sequence and sensor data. Our main research topic is on time and space efficient algorithms for pattern matching/search/discovery, data compression, and compressed data processing. We also study combinatorics on words as a theoretical background for algorithmics on strings.
Pattern matching/search/discovery
We study efficient algorithms and index structures for pattern matching and string information retrieval, used for example in keyword search, as well as finding patterns that characterize the data, based on various appropriate definitions of “occurrence”.
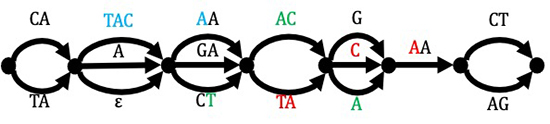
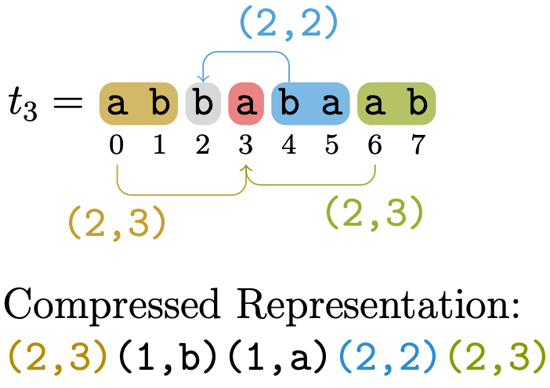
Data compression and compressed data processing
Through data compression, we aim not only to save storage space and communication bandwidth required for storing and transferring the data, but also to “compress” even the time required for computation, by processing the data “as is” in compressed form.
Software
Publications
Journal Papers
- Katsuhito Nakashima, Noriki Fujisato, Diptarama Hendrian, Yuto Nakashima, Ryo Yoshinaka, Shunsuke Inenaga, Hideo Bannai, Ayumi Shinohara, Masayuki Takeda, Parameterized DAWGs: Efficient constructions and bidirectional pattern searches, Theoretical Computer Science, 933:21-42, 2022.
https://doi.org/10.1016/j.tcs.2022.09.008 - Tooru Akagi, Yuki Kuhara, Takuya Mieno, Yuto Nakashima, Shunsuke Inenaga, Hideo Bannai, Masayuki Takeda, Combinatorics of minimal absent words for a sliding window, Theoretical Computer Science, 927: 109-119, 2022.
https://dx.doi.org/10.1016/j.tcs.2022.06.002 - Kazuya Tsuruta, Dominik Köppl, Shunsuke Kanda, Yuto Nakashima, Shunsuke Inenaga, Hideo Bannai, Masayuki Takeda, c-trie++: A Dynamic Trie Tailored for Fast Prefix Searches, Information and Computation, 285, Part B 104794, 2022.
https://doi.org/10.1016/j.ic.2021.104794 - Hiroe Inoue, Yoshiaki Matsuoka, Yuto Nakashima, Shunsuke Inenaga, Hideo Bannai, Masayuki Takeda, Factorizing Strings into Repetitions, Theory of Computing Systems, 66:484-501, 2022. https://dx.doi.org/10.1007/s00224-022-10070-3
- Takuya Mieno, Yuta Fujishige, Yuto Nakashima, Shunsuke Inenaga, Hideo Bannai, Masayuki Takeda, Computing Minimal Unique Substrings for a Sliding Window, Algorithmica, 84:670-693, 2022. https://dx.doi.org/10.1007/s00453-021-00864-1
- Takuya Mieno, Kiichi Watanabe, Yuto Nakashima, Shunsuke Inenaga, Hideo Bannai, Masayuki Takeda, Palindromic Trees for a Sliding Window and Its Applications, Information Processing Letters, 173:106174, 2022.
https://dx.doi.org/10.1016/j.ipl.2021.106174 - Dominik Köppl, Simon J. Puglisi, Rajeev Raman, Fast and Simple Compact Hashing via Bucketing, Algorithmica 84(9): 2735-2766, 2022.
https://doi.org/10.1007/s00453-022-00996-y - Dominik Köppl, Linking Off-Road Points to Routing Networks, Algorithms 15(5): 163, 2022. https://doi.org/10.3390/a15050163
- Dominik Köppl, Inferring Spatial Distance Rankings with Partial Knowledge on Routing Networks. Information 13(4): 168, 2022.
https://doi.org/10.3390/info13040168 - Paolo Ferragina, Giovanni Manzini, Travis Gagie, Dominik Köppl, Gonzalo Navarro, Manuel Striani, Francesco Tosoni, Improving Matrix-vector Multiplication via Lossless Grammar-Compressed Matrices, Proc. VLDB Endowment, 15(10): 2175-2187, 2022. https://doi.org/10.14778/3547305.3547321
- Alexandre P. Francisco, Travis Gagie, Dominik Köppl, Susana Ladra, Gonzalo Navarro, Graph Compression for Adjacency-Matrix Multiplication. SN Computer Science, 3(3): 193, 2022. https://doi.org/10.1007/s42979-022-01084-2
- Alexandre P. Francisco, Travis Gagie, Dominik Köppl, Susana Ladra, Gonzalo Navarro, Correction to: Graph Compression for Adjacency-Matrix Multiplication. SN Computer Science, 3(3): 228, 2022.
https://doi.org/10.1007/s42979-022-01141-w
Peer Reviewed Conference Proceedings
- Laurentius Leonard, Shunsuke Inenaga, Hideo Bannai, Takuya Mieno. Online algorithms for finding distinct substrings with length and multiple prefix and suffix conditions String Processing and Information Retrieval (SPIRE 2022). 2022.11;
- Hideo Bannai, Keisuke Goto, Masakazu Ishihata, Shunsuke Kanda, Dominik Köppl, Takaaki Nishimoto, Computing NP-hard Repetitiveness Measures via MAX-SAT, Proc. The 30th Annual European Symposium on Algorithms (ESA 2022), LIPIcs 244, 12:1-12:16, 2022. https://dx.doi.org/10.4230/LIPIcs.ESA.2022.12
- Tomohiro I, Dominik Köppl, Space-Efficient B Trees via Load-Balancing, Proc. International Workshop on Combinatorial Algorithms (IWOCA 2022), LNCS 13270: 327-340, 2022. https://doi.org/10.1007/978-3-031-06678-8_24
- Hideo Bannai, Tomohiro I, Tomasz Kociumaka, Dominik Köppl, Simon J. Puglisi. Computing Longest (Common) Lyndon Subsequences, Proc. International Workshop on Combinatorial Algorithms (IWOCA 2022), LNCS 13270: 128-142, 2022.
https://dx.doi.org/10.1007/978-3-031-06678-8_10 - Jin Jie Deng, Wing-Kai Hon, Dominik Köppl, Kunihiko Sadakane, FM-Indexing Grammars Induced by Suffix Sorting for Long Patterns, Proc. Data Compression Conference 2022 (DCC 2022): 63-72, 2022. https://doi.org/10.1109/DCC52660.2022.00014
- Dominik Köppl, Computing Lexicographic Parsings, Proc. Data Compression Conference 2022 (DCC 2022): 232-241, 2022.
https://doi.org/10.1109/DCC52660.2022.00031
Journal Papers
- Ryo Sugahara, Yuto Nakashima, Shunsuke Inenaga, Hideo Bannai, Masayuki Takeda. Efficiently computing runs on a trie, Theoretical Computer Science, 887:143-151, 2021. https://dx.doi.org/10.1016/j.tcs.2021.07.011
- Hideo Bannai, Shunsuke Inenaga, Neerja Mhaskar, Longest previous overlapping factor array Information Processing Letters, 168:106097, 2021. https://dx.doi.org/10.1016/j.ipl.2021.106097
- Shiori Mitsuya, Yuto Nakashima, Shunsuke Inenaga, Hideo Bannai, Masayuki Takeda, Compressed Communication Complexity of Hamming Distance, Algorithms, 14 (4): 116, 2021. https://dx.doi.org/10.3390/a14040116
- Mitsuru Funakoshi, Yuto Nakashima, Shunsuke Inenaga, Hideo Bannai, Masayuki Takeda, Computing longest palindromic substring after single-character or block-wise edits, Theoretical Computer Science. 2021.03; 859 116-133.
https://dx.doi.org/10.1016/j.tcs.2021.01.014 - Hideo Bannai, Momoko Hirayama, Danny Hucke, Shunsuke Inenaga, Artur Jez, Markus Lohrey, Carl Philipp Reh, The Smallest Grammar Problem Revisited IEEE Transactions on Information Theory, 67 (1): 317-328, 2021.
https://dx.doi.org/10.1109/tit.2020.3038147 - Dominik Köppl, Tomohiro I, Isamu Furuya, Yoshimasa Takabatake, Kensuke Sakai, Keisuke Goto, Re-Pair in Small Space, Algorithms 14(1): 5, 2021. https://doi.org/10.3390/a14010005
- Dominik Köppl, Non-Overlapping LZ77 Factorization and LZ78 Substring Compression Queries with Suffix Trees, Algorithms 14(2): 44, 2021. https://doi.org/10.3390/a14020044
- Dominik Köppl, Reversed Lempel-Ziv Factorization with Suffix Trees, Algorithms 14(6): 161, 2021. https://doi.org/10.3390/a14060161
- Diego Arroyuelo, Rodrigo Cánovas, Johannes Fischer, Dominik Köppl, Marvin Löbel, Gonzalo Navarro, Rajeev Raman, Engineering Practical Lempel-Ziv Tries. ACM Journal of Experimental Algorithmics 26:14:1-14:47, 2021. https://doi.org/10.1145/3481638
Peer Reviewed Conference Proceedings
- Tooru Akagi, Dominik Köppl, Yuto Nakashima, Shunsuke Inenaga, Hideo Bannai, Masayuki Takeda, Grammar Index by Induced Suffix Sorting, Proc. 28th International Symposium on String Processing and Information Retrieval (SPIRE 2021), LNCS 12944: 85-99, 2021. https://dx.doi.org/10.1007/978-3-030-86692-1_8
- Hideo Bannai, Mitsuru Funakoshi, Tomohiro I, Dominik Köppl, Takuya Mieno, Takaaki Nishimoto, A Separation of γ and b via Thue–Morse Words, Proc. 28th International Symposium on String Processing and Information Retrieval (SPIRE 2021), LNCS 12944:167-178, 2021. https://dx.doi.org/10.1007/978-3-030-86692-1_14
- Kosuke Fujita, Yuto Nakashima, Shunsuke Inenaga, Hideo Bannai, Masayuki Takeda. Longest Common Rollercoasters, Proc. 28th International Symposium on String Processing and Information Retrieval (SPIRE 2021), LNCS 12944:21-32.
https://dx.doi.org/10.1007/978-3-030-86692-1_3 - Tomohiro I, Robert W. Irving, Dominik Köppl, Lorna Love, Extracting the Sparse Longest Common Prefix Array from the Suffix Binary Search Tree, Proc. 28th International Symposium on String Processing and Information Retrieval (SPIRE 2021), LNCS 12944: 143-150, 2021. https://doi.org/10.1007/978-3-030-86692-1_12
- Hideo Bannai, Juha Kärkkäinen, Dominik Köppl, Marcin Piątkowski, Constructing the Bijective and the Extended Burrows-Wheeler Transform in Linear Time, Proc. 32nd Annual Symposium on Combinatorial Pattern Matching (CPM 2021), LIPIcs 191, 7:1-7:16, 2021. https://dx.doi.org/10.4230/LIPIcs.CPM.2021.7
- Noriki Fujisato, Yuto Nakashima, Shunsuke Inenaga, Hideo Bannai, Masayuki Takeda, The Parameterized Suffix Tray, Proc. 12th International Conference on Algorithms and Complexity (CIAC 2021), LNCS 12701: 258-270, 2021.
https://dx.doi.org/10.1007/978-3-030-75242-2_18 - Christina Boucher, Travis Gagie, Tomohiro I, Dominik Köppl, Ben Langmead, Giovanni Manzini, Gonzalo Navarro, Alejandro Pacheco, Massimiliano Rossi, PHONI: Streamed Matching Statistics with Multi-Genome References, Proc. Data Compression Conference 2021 (DCC 2021), 193-202, 2021. https://doi.org/10.1109/DCC50243.2021.00027
Journal Papers
- Takuya Mieno, Dominik Köppl, Yuto Nakashima, Shunsuke Inenaga, Hideo Bannai, Masayuki Takeda, Space-efficient algorithms for computing minimal/shortest unique substrings, Theoretical Computer Science, 845:230-242, 2020.
https://dx.doi.org/10.1016/j.tcs.2020.09.017 - Hideo Bannai, Travis Gagie, Gary Hoppenworth, Simon J. Puglisi, Luís M. S. Russo, More Time-Space Tradeoffs for Finding a Shortest Unique Substring, Algorithms, 13(9): 234, 2020. https://dx.doi.org/10.3390/a13090234
- Kazuya Tsuruta, Dominik Köppl, Yuto Nakashima, Shunsuke Inenaga, Hideo Bannai, Masayuki Takeda. Grammar-compressed Self-index with Lyndon Words, IPSJ Transactions on Mathematical Modeling and Its Applications, 13(2): 84-92, 2020.
http://id.nii.ac.jp/1001/00206600/
Peer Reviewed Conference Proceedings
- Akihiro Nishi, Yuto Nakashima, Shunsuke Inenaga, Hideo Bannai, Masayuki Takeda, Towards Efficient Interactive Computation of Dynamic Time Warping Distance, Proc. 27th International Symposium on String Processing and Information Retrieval (SPIRE 2020), LNCS 12303: 27-41, 2020. https://dx.doi.org/10.1007/978-3-030-59212-7_3
- Hideo Bannai, Takuya Mieno, Yuto Nakashima, Lyndon Words, the Three Squares Lemma, and Primitive Squares, Proc. 27th International Symposium on String Processing and Information Retrieval (SPIRE 2020), LNCS 12303: 265-273, 2020.
https://dx.doi.org/10.1007/978-3-030-59212-7_19 - Kanaru Kutsukake, Takuya Matsumoto, Yuto Nakashima, Shunsuke Inenaga, Hideo Bannai, Masayuki Takeda, On Repetitiveness Measures of Thue-Morse Words, Proc. 27th International Symposium on String Processing and Information Retrieval (SPIRE 2020), LNCS 12303: 213-220, 2020. https://dx.doi.org/10.1007/978-3-030-59212-7_15
- Takafumi Inoue, Shunsuke Inenaga, Hideo Bannai, Longest Square Subsequence Problem Revisited, Proc. 27th International Symposium on String Processing and Information Retrieval (SPIRE 2020), LNCS 12303: 147-154, 2020.
https://dx.doi.org/10.1007/978-3-030-59212-7_11
Journal Papers
- Yuto Nakashima, Takuya Takagi, Shunsuke Inenaga, Hideo Bannai, Masayuki Takeda, On the size of the smallest alphabet for Lyndon trees, Theoretical Computer Science, 792:131-143, 2019. https://doi.org/10.1016/j.tcs.2018.06.044
Peer Reviewed Conference Proceedings
- Yuta Fujishige, Yuto Nakashima, Shunsuke Inenaga, Hideo Bannai, Masayuki Takeda, An improved data structure for left-right maximal generic words problem, In Proc. 30th International Symposium on Algorithms and Computation (ISAAC 2019), LIPIcs 149, 40:1-40:12, 2019. https://doi.org/10.4230/LIPIcs.ISAAC.2019.40
- Noriki Fujisato, Yuto Nakashima, Shunsuke Inenaga, Hideo Bannai, Masayuki Takeda, Direct Linear Time Construction of Parameterized Suffix and LCP Arrays for Constant Alphabets, In Proc. 26th International Symposium on String Processing and Information Retrieval (SPIRE 2019), LNCS 11811:382-391, 2019. https://doi.org/10.1007/978-3-030-32686-9_27
- Kazuki Kai, Yuto Nakashima, Shunsuke Inenaga, Hideo Bannai, Masayuki Takeda, Tomasz Kociumaka, On Longest Common Property Preserved Substring Queries, In Proc. 26th International Symposium on String Processing and Information Retrieval (SPIRE 2019), LNCS 11811:162-174, 2019. https://doi.org/10.1007/978-3-030-32686-9_12
- Takuya Mieno, Dominik Koeppl, Yuto Nakashima, Shunsuke Inenaga, Hideo Bannai, Masayuki Takeda, Compact Data Structures for Shortest Unique Substring Queries, In Proc/ 26th International Symposium on String Processing and Information Retrieval (SPIRE 2019), LNCS 11811:107-123, 2019. https://doi.org/10.1007/978-3-030-32686-9_8
- Jarno Alanko, Hideo Bannai, Bastien Cazaux, Pierre Peterlongo, Jens Stoye, Finding all maximal perfect haplotype blocks in linear time, In Proc. 19th Workshop on Algorithms in Bioinformatics (WABI 2019), LIPIcs 143, 8:1-8:9, 2019. https://doi.org/10.4230/LIPIcs.WABI.2019.8
- Mitsuru Funakoshi, Yuto Nakashima, Shunsuke Inenaga, Hideo Bannai, Masayuki Takeda, Computing Maximal Palindromes and Distinct Palindromes in a Trie, In Proc. Prague Stringology Conference 2019 (PSC 2019), 3-15, 2019. https://www.stringology.org/event/2019/p02.html
- Golnaz Badkobeh, Hideo Bannai, Maxime Crochemore, Tomohiro I, Shunsuke Inenaga, Shiho Sugimoto, k-Abelian pattern matching: Revisited, corrected, and extended, In Proc. Prague Stringology Conference 2019 (PSC 2019), 29-40, 2019. https://www.stringology.org/event/2019/p04.html
- Kiichi Watanabe, Yuto Nakashima, Shunsuke Inenaga, Hideo Bannai, Masayuki Takeda, Shortest Unique Palindromic Substring Queries on Run-Length Encoded Strings, In Proc. 30th International Workshop on Combinatorial Algorithms (IWOCA 2019), LNCS 11638:430-441, 2019. https://doi.org/10.1007/978-3-030-25005-8_35
- Hideo Bannai, Juha Karkkainen, Dominik Koeppl, Marcin Piatkowski, Indexing the Bijective BWT, In Proc. 30th Annual Symposium on Combinatorial Pattern Matching (CPM 2019), LIPIcs 128, 17:1-17:14, 2019. https://doi.org/10.4230/LIPIcs.CPM.2019.17
- Ryo Sugahara, Yuto Nakashima, Shunsuke Inenaga, Hideo Bannai, Masayuki Takeda, Computing Runs on a Trie, In Proc. 30th Annual Symposium on Combinatorial Pattern Matching (CPM 2019), LIPIcs 128, 23:1-23:11, 2019. https://doi.org/10.4230/LIPIcs.CPM.2019.23
- Mitsuru Funakoshi, Yuto Nakashima, Shunsuke Inenaga, Hideo Bannai, Masayuki Takeda, Faster Queries for Longest Substring Palindrome After Block Edit, In Proc. 30th Annual Symposium on Combinatorial Pattern Matching (CPM 2019), LIPIcs 128, 27:1-27:13, 2019. https://doi.org/10.4230/LIPIcs.CPM.2019.27
- Yuki Urabe, Yuto Nakashima, Shunsuke Inenaga, Hideo Bannai, Masayuki Takeda, On the Size of Overlapping Lempel-Ziv and Lyndon Factorizations, In Proc. 30th Annual Symposium on Combinatorial Pattern Matching (CPM 2019), LIPIcs 128, 29:1-29:11, 2019. https://doi.org/10.4230/LIPIcs.CPM.2019.29
- Noriki Fujisato, Yuto Nakashima, Shunsuke Inenaga, Hideo Bannai, Masayuki Takeda, The Parameterized Position Heap of a Trie, In Proc. 11th International Conference on Algorithms and Complexity (CIAC 2019), LNCS 11485: 237-248, 2019. https://doi.org/10.1007/978-3-030-17402-6_20
- Isamu Furuya, Takuya Takagi, Yuto Nakashima, Shunsuke Inenaga, Hideo Bannai, Takuya Kida, MR-RePair: Grammar Compression Based on Maximal Repeats. In Proc. Data Compression Conference (DCC 2019), 508-517, 2019. https://doi.org/10.1109/DCC.2019.00059
Journal Papers
- Hideo Bannai, Travis Gagie, Shunsuke Inenaga, Juha Karkkainen, Dominik Kempa, Marcin Piatkowski, Shiho Sugimoto, Diverse Palindromic Factorization is NP-Complete, International Journal of Foundations of Computer Science. 29(2): 143-164, 2018. https://doi.org/10.1142/S0129054118400014
- Hiroe Inoue, Yuto Nakashima, Takuya Mieno, Shunsuke Inenaga, Hideo Bannai, Masayuki Takeda, Algorithms and combinatorial properties on shortest unique palindromic substrings, Journal of Discrete Algorithms 52-53: 122-132, 2018. https://doi.org/10.1016/j.jda.2018.11.009
Peer Reviewed Conference Proceedings
- Keisuke Goto, Tomohiro I, Hideo Bannai, Shunsuke Inenaga, Block Palindromes: A New Generalization of Palindromes, In Proc. 25th International Symposium on String Processing and Information Retrieval (SPIRE 2018), LNCS 11147: 183-190, 2018. https://doi.org/10.1007/978-3-030-00479-8_15
- Yuki Kuhara, Yuto Nakashima, Shunsuke Inenaga, Hideo Bannai, Masayuki Takeda, Recovering, Counting and Enumerating Strings from Forward and Backward Suffix Arrays, In Proc. 25th International Symposium on String Processing and Information Retrieval (SPIRE 2018), LNCS 11147: 254-267, 2018. https://doi.org/10.1007/978-3-030-00479-8_21
- Akihiro Nishi, Yuto Nakashima, Shunsuke Inenaga, Hideo Bannai, Masayuki Takeda, O(n log n)-time Text Compression by LZ-style Longest First Substitution, In Proc. Prague Stringology Conference 2018 (PSC 2018), 12-26, 2018. https://www.stringology.org/event/2018/p03.html
- Noriki Fujisato, Yuto Nakashima, Shunsuke Inenaga, Hideo Bannai Masayuki Takeda, Right-to-left Online Construction of Parameterized Position Heaps, In Proc. Prague Stringology Conference 2018 (PSC 2018), 91-102, 2018. https://www.stringology.org/event/2018/p09.html
- Rui Henriques, Alexandre Francisco, Luis Russo, Hideo Bannai, Order-Preserving Pattern Matching Indeterminate Strings, In Proc. 29th Annual Symposium on Combinatorial Pattern matching (CPM 2018), LIPIcs 105, 2:1-2:15, 2018. https://doi.org/10.4230/LIPIcs.CPM.2018.2
- Hideo Bannai, Travis Gagie, and Tomohiro I, Online LZ77 Parsing and Matching Statistics with RLBWTs, In Proc. 29th Annual Symposium on Combinatorial Pattern matching (CPM 2018), LIPIcs 105, 7:1-7:12, 2018. https://doi.org/10.4230/LIPIcs.CPM.2018.7
- Kotaro Aoyama, Yuto Nakashima, Tomohiro I, Shunsuke Inenaga, Hideo Bannai, Masayuki Takeda, Faster Online Elastic Degenerate String Matching, In Proc. 29th Annual Symposium on Combinatorial Pattern matching (CPM 2018), LIPIcs 105, 9:1-9:10, 2018. https://doi.org/10.4230/LIPIcs.CPM.2018.9
- Mitsuru Funakoshi, Yuto Nakashima, Shunsuke Inenaga, Hideo Bannai, Masayuki Takeda, Longest substring palindrome after edit, In Proc. 29th Annual Symposium on Combinatorial Pattern matching (CPM 2018), LIPIcs 105, 12:1-12:14, 2018. https://doi.org/10.4230/LIPIcs.CPM.2018.12
- Takafumi Inoue, Shunsuke Inenaga, Heikki Hyyro, Hideo Bannai, and Masayuki Takeda, Computing longest common square subsequences, In Proc. 29th Annual Symposium on Combinatorial Pattern matching (CPM 2018), LIPIcs 105, 15:1-15:13, 2018. https://doi.org/10.4230/LIPIcs.CPM.2018.15
- Yuki Urabe, Yuto Nakashima, Shunsuke Inenaga, Hideo Bannai, Masayuki Takeda, Longest Lyndon Substring After Edit, In Proc. 29th Annual Symposium on Combinatorial Pattern matching (CPM 2018), LIPIcs 105, 19:1-19:10, 2018. https://doi.org/10.4230/LIPIcs.CPM.2018.19
- Isamu Furuya, Yuto Nakashima, Tomohiro I, Shunsuke Inenaga, Hideo Bannai, Masayuki Takeda, Lyndon Factorization of Grammar Compressed Texts Revisited, In Proc. 29th Annual Symposium on Combinatorial Pattern matching (CPM 2018), LIPIcs 105, 24:1-24:10, 2018. https://doi.org/10.4230/LIPIcs.CPM.2018.24

Department of Integrated Analytics

Department of AI Technology Development

Department of Biostatistics
