統合解析分野
Department of Integrated Analytics
当分野では、生命科学・医歯学分野において急激に集積されているゲノムやエピゲノム、トランスクリプトーム、プロテオーム、メタボロームなどのオミクスデータとヒトの体組成値、問診項目、血液検査値、血液ガス分析値、心電図、人工呼吸器データなどが組み合わさった多次元・超ヘテロな生体ビッグデータを、生命情報学や統計科学、機械学習(ベイズ理論、深層学習など)とスーパーコンピュータやGPUなどの先端的情報処理技術を用いて解析することで、新たな医学・生命科学的知見を明らかにし、生命システムの解明に至ることを目標にしています。特に当分野では、既存の生命情報学的方法論の理解と適用に留まらず、続々と登場する新しい生命医科学データや多次元・超ヘテロな生体データ解析するための新しい数理的解析手法の開発までを行っています。これら新しい方法論と幅広い医学・生命科学への理解を通して、生体内システムの解析やゲノム・エピゲノムを原因とする病態の解明、急性・慢性期疾患を対象とした患者の予後や将来状態の予測、更には治療戦略の策定を実現します。
メンバー

宮野 悟
特任教授
長谷川 嵩矩
准教授
田中 洋子
助教
鈴木 麻子
技術補佐員
岡本 青史
(富士通株式会社 フェロー)
客員教授
丸橋 弘治
(富士通株式会社
富士通研究所人工知能研究所
プロジェクトマネージャ)
客員教授
研究プロジェクト
数理的モデリングを用いた生体内状態予測
疾患の病態は、生体内の遺伝子や代謝物、腸内や皮膚の細菌叢など、多次元・ヘテロな因子が複雑かつ相互に影響し合い、異常を引き起こしている状態です。一方で健常な状態とは、それらが生体内でバランスされている状態であるといえます。本研究室では、ゲノムやトランスクリプトーム、プロテオームなどのオミクスデータや臨床・健康診断を通して得られる大規模な患者データと生体内や病態の数理的モデリングを組み合わせることで、これらのデータ間の複雑な関係性を明らかにし、健常者や罹患者における生体内状態を明らかにする研究を行っています。これにより、疾患の原因となる因子の判別や、患者さん一人ひとりの重症化・予後予測、その治療戦略の提案が可能となります。またこれらの技術を応用することで、部分的な医療情報、例えば体組成値や問診項目だけが与えられたときに、血液検査値の予測を通して疾患の罹患確率を推定したり、生体内状態が異常となっている部位の判別などを行うことが可能になります。
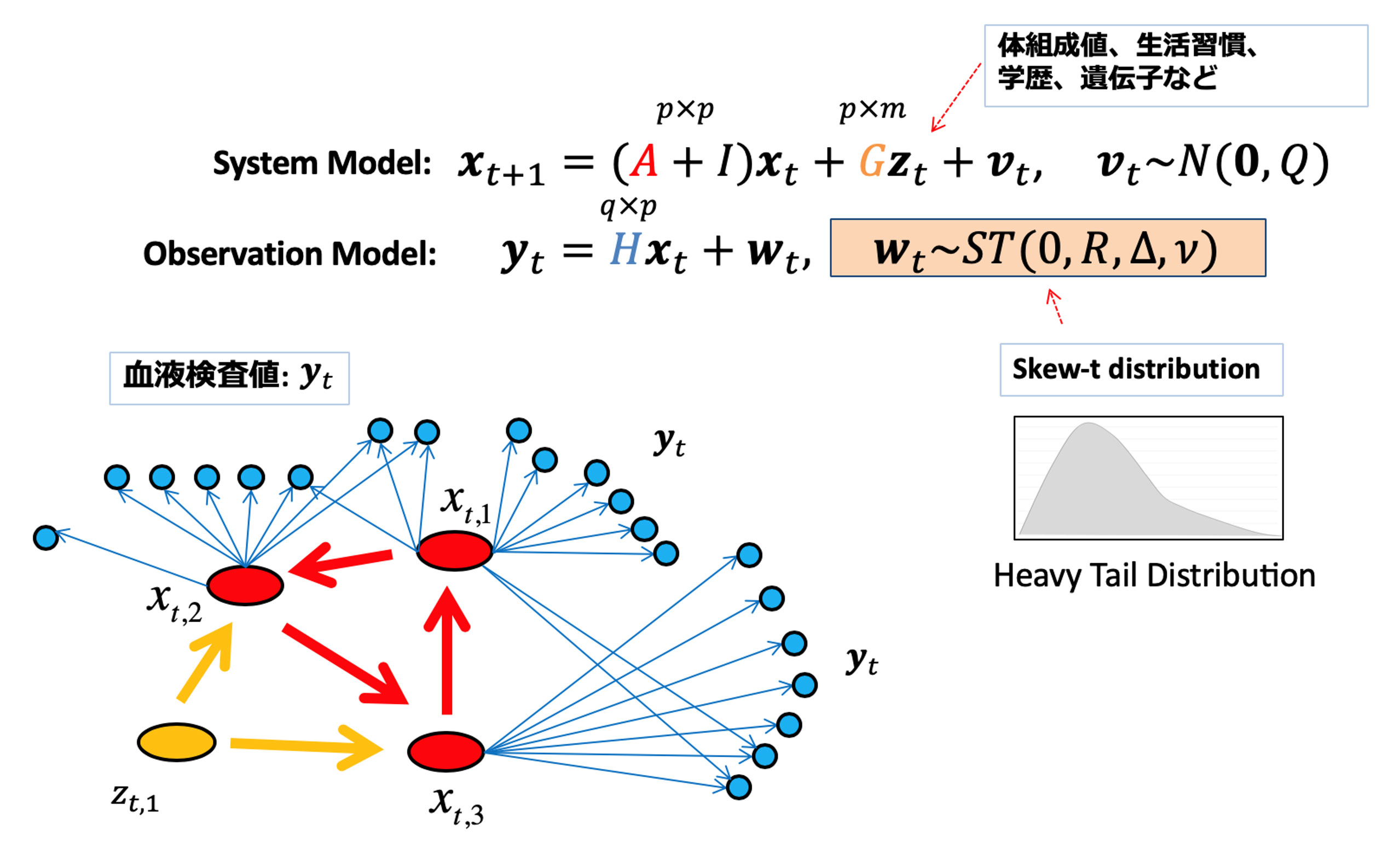
生活習慣と社会的ステータス、遺伝的要因が血液検査値に与える影響を状態空間モデルを用いて数理モデルを構築、健康状態の時系列シミュレーションを行う
腫瘍細胞や老化細胞などの炎症誘発細胞を標的とするためのネオアンチゲン予測手法開発
次世代シークエンサーの発展により、シークエンス解析は、がん研究において極めて標準的な解析手法となっています。当分野では長年の間、シークエンスの結果を数理的に解析することでがんの病態解明を明らかにする研究を行って来ました。その一つとして、ネオ抗原といわれる、近年活発に研究が行われている腫瘍特異的抗原に焦点を当てた研究を行なっており、同定された体細胞変異などの情報からネオ抗原を予測するための解析手法の開発やネオ抗原のがん免疫システムにおける役割の解析、腫瘍における免疫病態の評価を行っています。特に近年、老化細胞を標的とするように設計されたT細胞やチェックポイント阻害薬を用いることで、腫瘍細胞だけではなく、老化細胞を(部分的に)除去出来る可能性も示唆されており、研究が加速しています。我々は、免疫細胞を制御し、これらの細胞を適切に除去することを目的として、細胞レベルの免疫病態の評価と解明、また適切なネオ抗原予測を行う数理的解析手法を開発しています。
大規模データ解析と人工知能技術によるがんの起源と多様性の解明
長年のがん研究を通して、浸潤に関連する遺伝子やEMT(Epithelial Mesenchymal Transition:上皮間葉転換)に関連する遺伝子など、どの遺伝子ががんの転移に深く関与しているのかの特定が進められてきました。しかしながら、すべての細胞において、遺伝子同士は複雑な相互制御システムを形成しているため、浸潤やEMTのメカニズムを解明するには、単に関連している遺伝子を特定するだけでは不十分であり、関連している遺伝子同士がどのようなネットワークを形成しているのか、そのネットワーク構造の中で、特に浸潤やEMTに重要な役割を果たしているネットワークはどれなのか、そして、その司令塔となっている遺伝子はどれなのかなどを明らかにする必要があります。我々の研究チームは、東京大学医科学研究所ヒトゲノム解析センターのスーパーコンピュータやスーパーコンピュータ「京」・「富岳」を用い、ネットワークプロファイリングと呼ばれるネットワークを推定するプログラムを動かすことで、がん細胞の転移や再発に関連している可能性の高い遺伝子が形成しているネットワーク構造の抽出に取り組み、個々の遺伝子レベルではわからない転移や再発のメカニズムの解明や新たなメカニズムの発見を行っています。
M&Dデータの統合的解析基盤の構築と医歯学・病院との共同研究体制の推進
医歯学・医療研究は、情報の質と量双方の観点から、新たな時代に入っています。健康状態や疾患の解析において、ゲノム情報解析や1細胞解析を通して大量の生体情報を取得・利用するのは一般的になっており、それらの情報を統合的に解析することで、新たな知見や疾病の治療戦略を生み出すことが可能となりました。統合解析分野はこのような大規模データ解析を長年継続している研究チームで構成されており、東京医科歯科大学内のみならず、東京大学、神戸大学、九州大学など数多くの外部研究機関と連携し、生命医科学研究を推進しています。研究内容は幅広く、コロナ制圧タスクフォースにおける全ゲノム解析の実施、ICUにおけるCOVID19患者の重症化予測を含む各種急性期疾患の重症化・死亡・予後予測研究、健常人・疾病・腫瘍ゲノム解析(全ゲノム、エクソーム、トランスクリプトーム、シングルセル解析、cfDNA解析、プロテオーム解析、メタボローム解析など)、薬剤の応答予測などを実施しています。加えて、東京大学医科学研究所のスーパーコンピュータSHIROKANEとの包括契約など、既設の組織と有機的に連携することで、本学の医療ビッグデータの統合的解析を実践すると共に、統合的解析に必要な情報教育と情報基盤の構築を推進しています。
研究内容
- 病態の数理的モデリングを用いた生体内状態予測
- 健康診断や診療データを用いた疾患の罹患確率、重症度予測の研究
- 数理モデリングとAIに基づく腫瘍免疫システムの解明
- スーパーコンピュータと数理モデリングを用いたがんの起源と多様性の解明
- 健常人・疾病・腫瘍を対象とするマルチオミクス大規模データ解析の研究
最近の主な研究業績
- Saiki R, Momozawa Y, Nannya Y, Nakagawa MM, Ochi Y, Yoshizato T, Terao C, Kuroda Y, Shiraishi Y, Chiba K, Tanaka H, Niida A, Imoto S, Matsuda K, Morisaki T, Murakami Y, Kamatani Y, Matsuda S, Kubo M, Miyano S, Makishima H, Ogawa S. Combined landscape of single-nucleotide variants and copy number alterations in clonal hematopoiesis. Nature medicine. 2021.07; 27 (7): 1239-1249. ( PubMed, DOI )
- Fujii Y, Sato Y, Suzuki H, Kakiuchi N, Yoshizato T, Lenis AT, Maekawa S, Yokoyama A, Takeuchi Y, Inoue Y, Ochi Y, Shiozawa Y, Aoki K, Yoshida K, Kataoka K, Nakagawa MM, Nannya Y, Makishima H, Miyakawa J, Kawai T, Morikawa T, Shiraishi Y, Chiba K, Tanaka H, Nagae G, Sanada M, Sugihara E, Sato TA, Nakagawa T, Fukayama M, Ushiku T, Aburatani H, Miyano S, Coleman JA, Homma Y, Solit DB, Kume H, Ogawa S. Molecular classification and diagnostics of upper urinary tract urothelial carcinoma. Cancer cell. 2021.06; 39 (6): 793-809.e8. ( PubMed, DOI )
- Yamaguchi K, Kasajima R, Takane K, Hatakeyama S, Shimizu E, Yamaguchi R, Katayama K, Arai M, Ishioka C, Iwama T, Kaneko S, Matsubara N, Moriya Y, Nomizu T, Sugano K, Tamura K, Tomita N, Yoshida T, Sugihara K, Nakamura Y, Miyano S, Imoto S, Furukawa Y, Ikenoue T. Application of targeted nanopore sequencing for the screening and determination of structural variants in patients with Lynch syndrome. Journal of human genetics. 2021.05; ( PubMed, DOI )
- Niida A, Mimori K, Shibata T, Miyano S. Modeling colorectal cancer evolution. Journal of human genetics. 2021.05; ( PubMed, DOI )
- Ochi Y, Yoshida K, Huang YJ, Kuo MC, Nannya Y, Sasaki K, Mitani K, Hosoya N, Hiramoto N, Ishikawa T, Branford S, Shanmuganathan N, Ohyashiki K, Takahashi N, Takaku T, Tsuchiya S, Kanemura N, Nakamura N, Ueda Y, Yoshihara S, Bera R, Shiozawa Y, Zhao L, Takeda J, Watatani Y, Okuda R, Makishima H, Shiraishi Y, Chiba K, Tanaka H, Sanada M, Takaori-Kondo A, Miyano S, Ogawa S, Shih LY. Clonal evolution and clinical implications of genetic abnormalities in blastic transformation of chronic myeloid leukaemia. Nature communications. 2021.05; 12 (1): 2833. ( PubMed, DOI )
- Yoshihiro Ishida, Nobuyuki Kakiuchi, Kenichi Yoshida, Yoshikage Inoue, Hiroyuki Irie, Tatsuki R Kataoka, Masahiro Hirata, Takeru Funakoshi, Shigeto Matsushita, Hiroo Hata, Hiroshi Uchi, Yuki Yamamoto, Yasuhiro Fujisawa, Taku Fujimura, Ryunosuke Saiki, Kengo Takeuchi, Yuichi Shiraishi, Kenichi Chiba, Hiroko Tanaka, Atsushi Otsuka, Satoru Miyano, Kenji Kabashima, Seishi Ogawa. Unbiased Detection of Driver Mutations in Extramammary Paget Disease. Clin Cancer Res. 2021.03; 27 (6): 1756-1765. ( PubMed, DOI )
- Kazuyuki Shimada, Kenichi Yoshida, Yasuhiro Suzuki, Chisako Iriyama, Yoshikage Inoue, Masashi Sanada, Keisuke Kataoka, Masaaki Yuge, Yusuke Takagi, Shigeru Kusumoto, Yasufumi Masaki, Takahiko Ito, Yuichiro Inagaki, Akinao Okamoto, Yachiyo Kuwatsuka, Masahiro Nakatochi, Satoko Shimada, Hiroaki Miyoshi, Yuichi Shiraishi, Kenichi Chiba, Hiroko Tanaka, Satoru Miyano, Yusuke Shiozawa, Yasuhito Nannya, Asako Okabe, Kei Kohno, Yoshiko Atsuta, Koichi Ohshima, Shigeo Nakamura, Seishi Ogawa, Akihiro Tomita, Hitoshi Kiyoi. Frequent genetic alterations in immune checkpoint-related genes in intravascular large B-cell lymphoma. Blood. 2021.03; 137 (11): 1491-1502. ( PubMed, DOI )
- Chen Li, Jiale Qin, Keisuke Kuroyanagi, Lu Lu, Masao Nagasaki, Miyano Satoru. High-speed parameter search of dynamic biological pathways from time-course transcriptomic profiles using high-level Petri net. Biosystems. 2021.03; 201 104332. ( PubMed, DOI )
- Taeko Kaburagi, Genki Yamato, Norio Shiba, Kenichi Yoshida, Yusuke Hara, Ken Tabuchi, Yuichi Shiraishi, Kentaro Ohki, Manabu Sotomatsu, Hirokazu Arakawa, Hidemasa Matsuo, Akira Shimada, Tomohiko Taki, Nobutaka Kiyokawa, Daisuke Tomizawa, Keizo Horibe, Satoru Miyano, Takashi Taga, Souichi Adachi, Seishi Ogawa, Yasuhide Hayashi. Clinical significance of RAS pathway alterations in pediatric acute myeloid leukemia. Haematologica. 2021.03; Online ahead of print ( PubMed, DOI )
- Suzuki M, Kasajima R, Yokose T, Ito H, Shimizu E, Hatakeyama S, Yokoyama K, Yamaguchi R, Furukawa Y, Miyano S, Imoto S, Yoshioka E, Washimi K, Okubo Y, Kawachi K, Sato S, Miyagi Y. Comprehensive molecular analysis of genomic profiles and PD-L1 expression in lung adenocarcinoma with a high-grade fetal adenocarcinoma component. Translational lung cancer research. 2021.03; 10 (3): 1292-1304. ( PubMed, DOI )
- Akira Nishimura, Shinsuke Hirabayashi, Daisuke Hasegawa, Kenichi Yoshida, Yuichi Shiraishi, Miho Ashiarai, Yosuke Hosoya, Tohru Fujiwara, Hideo Harigae, Satoru Miyano, Seishi Ogawa, Atsushi Manabe. Acquisition of monosomy 7 and a RUNX1 mutation in Pearson syndrome. Pediatr Blood Cancer. 2021.02; 68 (2): e28799. ( PubMed, DOI )
- Nobutaka Ebata, Masashi Fujita, Shota Sasagawa, Kazuhiro Maejima, Yuki Okawa, Yutaka Hatanaka, Tomoko Mitsuhashi, Ayako Oosawa-Tatsuguchi, Hiroko Tanaka, Satoru Miyano, Toru Nakamura, Satoshi Hirano, Hidewaki Nakagawa. Molecular Classification and Tumor Microenvironment Characterization of Gallbladder Cancer by Comprehensive Genomic and Transcriptomic Analysis. Cancers (Basel). 2021.02; 13 (4): 733. ( PubMed, DOI )
- Kosuke Fujimoto, Yasumasa Kimura, Jessica R Allegretti, Mako Yamamoto, Yao-Zhong Zhang, Kotoe Katayama, Georg Tremmel, Yunosuke Kawaguchi, Masaki Shimohigoshi, Tetsuya Hayashi, Miho Uematsu, Kiyoshi Yamaguchi, Yoichi Furukawa, Yutaka Akiyama, Rui Yamaguchi, Sheila E Crowe, Peter B Ernst, Satoru Miyano, Hiroshi Kiyono, Seiya Imoto, Satoshi Uematsu. Functional Restoration of Bacteriomes and Viromes by Fecal Microbiota Transplantation. Gastroenterology. 2021.02; S0016-5085(21)00400-5 ( PubMed, DOI )
- Chantana Polprasert, Yasuhide Takeuchi, Hideki Makishima, Kitsada Wudhikarn, Nobuyuki Kakiuchi, Nichthida Tangnuntachai, Thamathorn Assanasen, Wimonmas Sitthi, Hamidah Muhamad, Panisinee Lawasut, Sunisa Kongkiatkamon, Udomsak Bunworasate, Koji Izutsu, Yuichi Shiraishi, Kenichi Chiba, Hiroko Tanaka, Satoru Miyano, Seishi Ogawa, Kenichi Yoshida, Ponlapat Rojnuckarin. Frequent mutations in HLA and related genes in extranodal NK/T cell lymphomas. Leuk Lymphoma. 2021.01; 62 (1): 95-103. ( PubMed, DOI )
- Heewon Park, Koji Maruhashi, Rui Yamaguchi, Seiya Imoto, Satoru Miyano. Correction: Global gene network exploration based on explainable artificial intelligence approach. PLoS One. 2021.01; 16 (1): e0246380. ( PubMed, DOI )
- Fumi Nakamura, Honoka Arai, Yasuhito Nannya, Motoshi Ichikawa, Shiho Furuichi, Fusako Nagasawa, Wataru Takahashi, Tomoyuki Handa, Yuko Nakamura, Hiroko Tanaka, Yuka Nakamura, Ko Sasaki, Satoru Miyano, Seishi Ogawa, Kinuko Mitani. Development of Philadelphia chromosome-negative acute myeloid leukemia with IDH2 and NPM1 mutations in a patient with chronic myeloid leukemia who showed a major molecular response to tyrosine kinase inhibitor therapy. Int J Hematol. 2021.01; Online ahead of print ( PubMed, DOI )
- Shunsuke Kimura, Masahiro Sekiguchi, Kentaro Watanabe, Mitsuteru Hiwatarai, Masafumi Seki, Kenichi Yoshida, Tomoya Isobe, Yusuke Shiozawa, Hiromichi Suzuki, Noriko Hoshino, Yasuhide Hayashi, Akira Oka, Satoru Miyano, Seishi Ogawa, Junko Takita. Association of high-risk neuroblastoma classification based on expression profiles with differentiation and metabolism. PLoS One. 2021.01; 16 (1): e0245526. ( PubMed, DOI )
- Shin-Ei Kudo, Yuta Kouyama, Yushi Ogawa, Katsuro Ichimasa, Tsuyoshi Hamada, Kazuki Kato, Koki Kudo, Takaaki Masuda, Hajime Otsu, Masashi Misawa, Yuichi Mori, Toyoki Kudo, Takemasa Hayashi, Kunihiko Wakamura, Hideyuki Miyachi, Naruhiko Sawada, Toshiro Sato, Tatsuhiro Shibata, Shigeharu Hamatani, Tetsuo Nemoto, Fumio Ishida, Atsushi Niida, Satoru Miyano, Masanobu Oshima, Shuji Ogino, Koshi Mimori. Depressed Colorectal Cancer: A New Paradigm in Early Colorectal Cancer. Clin Transl Gastroenterol. 2020.12; 11 (12): e00269. ( PubMed, DOI )
- Felix A Dingler, Meng Wang, Anfeng Mu, Christopher L Millington, Nina Oberbeck, Sam Watcham, Lucas B Pontel, Ashley N Kamimae-Lanning, Frederic Langevin, Camille Nadler, Rebecca L Cordell, Paul S Monks, Rui Yu, Nicola K Wilson, Asuka Hira, Kenichi Yoshida, Minako Mori, Yusuke Okamoto, Yusuke Okuno, Hideki Muramatsu, Yuichi Shiraishi, Masayuki Kobayashi, Toshinori Moriguchi, Tomoo Osumi, Motohiro Kato, Satoru Miyano, Etsuro Ito, Seiji Kojima, Hiromasa Yabe, Miharu Yabe, Keitaro Matsuo, Seishi Ogawa, Berthold Gottgens, Michael R G Hodskinson, Minoru Takata, Ketan J Patel. Two Aldehyde Clearance Systems Are Essential to Prevent Lethal Formaldehyde Accumulation in Mice and Humans. Mol Cell. 2020.12; 80 (6): 996-1012.e9.
- Takashi Kanamori, Masashi Sanada, Masaki Ri, Hiroo Ueno, Dai Nishijima, Takahiko Yasuda, Takuto Tachita, Tomoko Narita, Shigeru Kusumoto, Atsushi Inagaki, Rei Ishihara, Yuki Murakami, Nobuhiko Kobayashi, Yusuke Shiozawa, Kenichi Yoshida, Masahiro M Nakagawa, Yasuhito Nannya, Yuichi Shiraishi, Kenichi Chiba, Hiroko Tanaka, Satoru Miyano, Keizo Horibe, Hiroshi Handa, Seishi Ogawa, Shinsuke Iida. Genomic analysis of multiple myeloma using targeted capture sequencing in the Japanese cohort. Br J Haematol. 2020.12; 191 (5): 755-763. ( PubMed, DOI )
- Heewon Park, Rui Yamaguchi, Seiya Imoto, Satoru Miyano. Automatic sparse principal component analysis The Canadian J Statistics. 2020.12; Online ahead of print ( DOI )
- Felix A Dingler, Meng Wang, Anfeng Mu, Christopher L Millington, Nina Oberbeck, Sam Watcham, Lucas B Pontel, Ashley N Kamimae-Lanning, Frederic Langevin, Camille Nadler, Rebecca L Cordell, Paul S Monks, Rui Yu, Nicola K Wilson, Asuka Hira, Kenichi Yoshida, Minako Mori, Yusuke Okamoto, Yusuke Okuno, Hideki Muramatsu, Yuichi Shiraishi, Masayuki Kobayashi, Toshinori Moriguchi, Tomoo Osumi, Motohiro Kato, Satoru Miyano, Etsuro Ito, Seiji Kojima, Hiromasa Yabe, Miharu Yabe, Keitaro Matsuo, Seishi Ogawa, Berthold Gottgens, Michael R G Hodskinson, Minoru Takata, Ketan J Patel. Two Aldehyde Clearance Systems Are Essential to Prevent Lethal Formaldehyde Accumulation in Mice and Humans. Mol Cell. 2020.12; 80 (6): 996-1012.e9. ( PubMed, DOI )
- Heewon Park, Koji Maruhashi, Rui Yamaguchi, Seiya Imoto, Satoru Miyano. Global gene network exploration based on explainable artificial intelligence approach. PLoS One. 2020.11; 15 (11): e0241508. ( PubMed, DOI )
- Sachiko Ishida, Kumiko Kato, Masami Tanaka, Toshitaka Odamaki, Ryuichi Kubo, Eri Mitsuyama, Jin-Zhong Xiao, Rui Yamaguchi, Satoshi Uematsu, Seiya Imoto, Satoru Miyano. Genome-wide association studies and heritability analysis reveal the involvement of host genetics in the Japanese gut microbiota. Commun Biol. 2020.11; 3 (1): 686. ( PubMed, DOI )
- Hiromi Ogura, Shouichi Ohga, Takako Aoki, Taiju Utsugisawa, Hidehiro Takahashi, Asayuki Iwai, Kenichiro Watanabe, Yusuke Okuno, Kenichi Yoshida, Seishi Ogawa, Satoru Miyano, Seiji Kojima, Toshiyuki Yamamoto, Keiko Yamamoto-Shimojima, Hitoshi Kanno. Novel COL4A1 mutations identified in infants with congenital hemolytic anemia in association with brain malformations. Hum Genome Var. 2020.11; 7 (1): 42. ( PubMed, DOI )
- Matthew H Bailey, William U Meyerson, Lewis Jonathan Dursi, Liang-Bo Wang, Guanlan Dong, Wen-Wei Liang, Amila Weerasinghe, Shantao Li, Yize Li, Sean Kelso, Gordon Saksena, Kyle Ellrott, Michael C Wendl, David A Wheeler, Gad Getz, Jared T Simpson, Mark B Gerstein, Li Ding, PCAWG Consortium (Satoru Miyano, et al.). Author Correction: Retrospective evaluation of whole exome and genome mutation calls in 746 cancer samples. Nat Commun. 2020.11; 11 (1): 6232. ( PubMed, DOI )
- Hidemasa Matsuo, Kenichi Yoshida, Kana Nakatani, Yutarou Harata, Moe Higashitani, Yuri Ito, Yasuhiko Kamikubo, Yusuke Shiozawa, Yuichi Shiraishi, Kenichi Chiba, Hiroko Tanaka, Ai Okada, Yasuhito Nannya, June Takeda, Hiroo Ueno, Nobutaka Kiyokawa, Daisuke Tomizawa, Takashi Taga, Akio Tawa, Satoru Miyano, Manja Meggendorfer, Claudia Haferlach, Seishi Ogawa, Souichi Adachi. Fusion partner-specific mutation profiles and KRAS mutations as adverse prognostic factors in MLL-rearranged AML. Blood Adv. 2020.10; 4 (19): 4623-4631. ( PubMed, DOI )
- Yukiko Inagaki-Kawata, Kenichi Yoshida, Nobuko Kawaguchi-Sakita, Masahiro Kawashima, Tomomi Nishimura, Noriko Senda, Yusuke Shiozawa, Yasuhide Takeuchi, Yoshikage Inoue, Aiko Sato-Otsubo, Yoichi Fujii, Yasuhito Nannya, Eiji Suzuki, Masahiro Takada, Hiroko Tanaka, Yuichi Shiraishi, Kenichi Chiba, Yuki Kataoka, Masae Torii, Hiroshi Yoshibayashi, Kazuhiko Yamagami, Ryuji Okamura, Yoshio Moriguchi, Hironori Kato, Shigeru Tsuyuki, Akira Yamauchi, Hirofumi Suwa, Takashi Inamoto, Satoru Miyano, Seishi Ogawa, Masakazu Toi. Genetic and clinical landscape of breast cancers with germline BRCA1/2 variants. Commun Biol. 2020.10; 3 (1): 578. ( PubMed, DOI )
- Hiroo Ueno, Kenichi Yoshida, Yusuke Shiozawa, Yasuhito Nannya, Yuka Iijima-Yamashita, Nobutaka Kiyokawa, Yuichi Shiraishi, Kenichi Chiba, Hiroko Tanaka, Tomoya Isobe, Masafumi Seki, Shunsuke Kimura, Hideki Makishima, Masahiro M Nakagawa, Nobuyuki Kakiuchi, Keisuke Kataoka, Tetsuichi Yoshizato, Dai Nishijima, Takao Deguchi, Kentaro Ohki, Atsushi Sato, Hiroyuki Takahashi, Yoshiko Hashii, Sadao Tokimasa, Junichi Hara, Yoshiyuki Kosaka, Koji Kato, Takeshi Inukai, Junko Takita, Toshihiko Imamura, Satoru Miyano, Atsushi Manabe, Keizo Horibe, Seishi Ogawa, Masashi Sanada. Landscape of driver mutations and their clinical impacts in pediatric B-cell precursor acute lymphoblastic leukemia. Blood Adv. 2020.10; 4 (20): 5165-5173. ( PubMed, DOI )
- Shun Koyamaishi, Takuya Kamio, Akie Kobayashi, Tomohiko Sato, Ko Kudo, Shinya Sasaki, Rika Kanezaki, Daiichiro Hasegawa, Hideki Muramatsu, Yoshiyuki Takahashi, Yoji Sasahara, Hidefumi Hiramatsu, Harumi Kakuda, Miyuki Tanaka, Masataka Ishimura, Masanori Nishi, Akira Ishiguro, Hiromasa Yabe, Takeo Sarashina, Masaki Yamamoto, Yuki Yuza, Nobuyuki Hyakuna, Kenichi Yoshida, Hitoshi Kanno, Shouichi Ohga, Akira Ohara, Seiji Kojima, Satoru Miyano, Seishi Ogawa, Tsutomu Toki, Kiminori Terui, Etsuro Ito. Correction: Reduced-intensity conditioning is effective for hematopoietic stem cell transplantation in young pediatric patients with Diamond-Blackfan anemia. Bone Marrow Transplant. 2020.10; Online ahead of print ( PubMed, DOI )
- Takuya Moriyama, Seiya Imoto, Satoru Miyano, Rui Yamaguchi. Theoretical Foundation of the Performance of Phylogeny-Based Somatic Variant Detection Lecture Note in Bioinformatis. 2020.10; 12508 87-101. ( DOI )
- 奥田 瑠璃花, 南谷 泰仁, 越智 陽太郎, 蝶名林 和久, Creignou Maria, 牧島 秀樹, 竹田 淳恵, 昆 彩奈, 宮野 悟, 半田 寛, 千葉 滋, 大屋敷 一馬, Haferlach Torsten, Lindberg Eva, 小川 誠司. 骨髄異形成症候群におけるder(1;7)の民族的、臨床的、遺伝的特徴(Distinct ethnic, clinical, and genetic characteristics of der(1;7) in myelodysplastic syndromes) 日本癌学会総会記事. 2020.10; 79回 OE7-1. ( 医中誌 )
- Takanori Hasegawa, Shuto Hayashi, Eigo Shimizu, Shinichi Mizuno, Atsushi Niida, Rui Yamaguchi, Satoru Miyano, Hidewaki Nakagawa, Seiya Imoto. Neoantimon: a multifunctional R package for identification of tumor-specific neoantigens. Bioinformatics. 2020.09; 36 (18): 4813-4816. ( PubMed, DOI )
- Shun Koyamaishi, Takuya Kamio, Akie Kobayashi, Tomohiko Sato, Ko Kudo, Shinya Sasaki, Rika Kanezaki, Daiichiro Hasegawa, Hideki Muramatsu, Yoshiyuki Takahashi, Yoji Sasahara, Hidefumi Hiramatsu, Harumi Kakuda, Miyuki Tanaka, Masataka Ishimura, Masanori Nishi, Akira Ishiguro, Hiromasa Yabe, Takeo Sarashina, Masaki Yamamoto, Yuki Yuza, Nobuyuki Hyakuna, Kenichi Yoshida, Hitoshi Kanno, Shouichi Ohga, Akira Ohara, Seiji Kojima, Satoru Miyano, Seishi Ogawa, Tsutomu Toki, Kiminori Terui, Etsuro Ito. Reduced-intensity conditioning is effective for hematopoietic stem cell transplantation in young pediatric patients with Diamond-Blackfan anemia. Bone Marrow Transplant. 2020.09; ( PubMed, DOI )
- Yasuo Kubota, Masafumi Seki, Tomoko Kawai, Tomoya Isobe, Misa Yoshida, Masahiro Sekiguchi, Shunsuke Kimura, Kentaro Watanabe, Aiko Sato-Otsubo, Kenichi Yoshida, Hiromichi Suzuki, Keisuke Kataoka, Yoichi Fujii, Yuichi Shiraishi, Kenichi Chiba, Hiroko Tanaka, Mitsuteru Hiwatari, Akira Oka, Yasuhide Hayashi, Satoru Miyano, Seishi Ogawa, Kenichiro Hata, Yukichi Tanaka, Junko Takita. Comprehensive genetic analysis of pediatric germ cell tumors identifies potential drug targets. Commun Biol. 2020.09; 3 (1): 544. ( PubMed, DOI )
- Kosuke Fujimoto, Yasumasa Kimura, Masaki Shimohigoshi, Takeshi Satoh, Shintaro Sato, Georg Tremmel, Miho Uematsu, Yunosuke Kawaguchi, Yuki Usui, Yoshiko Nakano, Tetsuya Hayashi, Koji Kashima, Yoshikazu Yuki, Kiyoshi Yamaguchi, Yoichi Furukawa, Masanori Kakuta, Yutaka Akiyama, Rui Yamaguchi, Sheila E Crowe, Peter B Ernst, Satoru Miyano, Hiroshi Kiyono, Seiya Imoto, Satoshi Uematsu. Metagenome Data on Intestinal Phage-Bacteria Associations Aids the Development of Phage Therapy against Pathobionts. Cell Host Microbe. 2020.09; 28 (3): 380-389.e9. ( PubMed, DOI )
- Matthew H Bailey, William U Meyerson, Lewis Jonathan Dursi, Liang-Bo Wang, Guanlan Dong, Wen-Wei Liang, Amila Weerasinghe, Shantao Li, Yize Li, Sean Kelso, , , Gordon Saksena, Kyle Ellrott, Michael C Wendl, David A Wheeler, Gad Getz, Jared T Simpson, Mark B Gerstein, Li Ding, . Retrospective evaluation of whole exome and genome mutation calls in 746 cancer samples. Nat Commun. 2020.09; 11 (1): 4748. ( PubMed, DOI )
- Constance H Li, Stephenie D Prokopec, Ren X Sun, Fouad Yousif, Nathaniel Schmitz, Paul C Boutros, PCAWG Consortium (Satoru Miyano, et al.). Sex differences in oncogenic mutational processes. Nat Commun. 2020.08; 11 (1): 4330. ( PubMed, DOI )
- Masahiro Sekiguchi, Masafumi Seki, Tomoko Kawai, Kenichi Yoshida, Misa Yoshida, Tomoya Isobe, Noriko Hoshino, Ryota Shirai, Mio Tanaka, Ryota Souzaki, Kentaro Watanabe, Yuki Arakawa, Yasuhito Nannya, Hiromichi Suzuki, Yoichi Fujii, Keisuke Kataoka, Yuichi Shiraishi, Kenichi Chiba, Hiroko Tanaka, Teppei Shimamura, Yusuke Sato, Aiko Sato-Otsubo, Shunsuke Kimura, Yasuo Kubota, Mitsuteru Hiwatari, Katsuyoshi Koh, Yasuhide Hayashi, Yutaka Kanamori, Mureo Kasahara, Kenichi Kohashi, Motohiro Kato, Takako Yoshioka, Kimikazu Matsumoto, Akira Oka, Tomoaki Taguchi, Masashi Sanada, Yukichi Tanaka, Satoru Miyano, Kenichiro Hata, Seishi Ogawa, Junko Takita. Integrated multiomics analysis of hepatoblastoma unravels its heterogeneity and provides novel druggable targets. NPJ Precis Oncol. 2020.07; 4 20. ( PubMed, DOI )
- Munmee Dutta, Hidewaki Nakagawa, Hiroaki Kato, Kazuhiro Maejima, Shota Sasagawa, Kaoru Nakano, Aya Sasaki-Oku, Akihiro Fujimoto, Raul Nicolas Mateos, Ashwini Patil, Hiroko Tanaka, Satoru Miyano, Takushi Yasuda, Kenta Nakai, Masashi Fujita. Whole genome sequencing analysis identifies recurrent structural alterations in esophageal squamous cell carcinoma. PeerJ. 2020.06; 8 e9294. ( PubMed, DOI )
- Yotaro Ochi, Ayana Kon, Toyonori Sakata, Masahiro M Nakagawa, Naotaka Nakazawa, Masanori Kakuta, Keisuke Kataoka, Haruhiko Koseki, Manabu Nakayama, Daisuke Morishita, Tatsuaki Tsuruyama, Ryunosuke Saiki, Akinori Yoda, Rurika Okuda, Tetsuichi Yoshizato, Kenichi Yoshida, Yusuke Shiozawa, Yasuhito Nannya, Shinichi Kotani, Yasunori Kogure, Nobuyuki Kakiuchi, Tomomi Nishimura, Hideki Makishima, Luca Malcovati, Akihiko Yokoyama, Kengo Takeuchi, Eiji Sugihara, Taka-Aki Sato, Masashi Sanada, Akifumi Takaori-Kondo, Mario Cazzola, Mineko Kengaku, Satoru Miyano, Katsuhiko Shirahige, Hiroshi I Suzuki, Seishi Ogawa. Combined Cohesin-RUNX1 Deficiency Synergistically Perturbs Chromatin Looping and Causes Myelodysplastic Syndromes. Cancer Discov. 2020.06; 10 (6): 836-853. ( PubMed, DOI )
- Yuki Saito, Junji Koya, Mitsugu Araki, Yasunori Kogure, Sumito Shingaki, Mariko Tabata, Marni B McClure, Kota Yoshifuji, Shigeyuki Matsumoto, Yuta Isaka, Hiroko Tanaka, Takanori Kanai, Satoru Miyano, Yuichi Shiraishi, Yasushi Okuno, Keisuke Kataoka. Landscape and function of multiple mutations within individual oncogenes. Nature. 2020.06; 582 (7810): 95-99. ( PubMed, DOI )
- Isidro Cortes-Ciriano, Jake June-Koo Lee, Ruibin Xi, Dhawal Jain, Youngsook L Jung, Lixing Yang, Dmitry Gordenin, Leszek J Klimczak, Cheng-Zhong Zhang, David S Pellman, Peter J Park, PCAWG Consortium (Satoru Miyano, et al.). Publisher Correction: Comprehensive analysis of chromothripsis in 2,658 human cancers using whole-genome sequencing. Nat Genet. 2020.05; Online ahead of print ( PubMed, DOI )
- Minako Mori, Asuka Hira, Kenichi Yoshida, Hideki Muramatsu, Yusuke Okuno, Yuichi Shiraishi, Michiko Anmae, Jun Yasuda, Shu Tadaka, Kengo Kinoshita, Tomoo Osumi, Yasushi Noguchi, Souichi Adachi, Ryoji Kobayashi, Hiroshi Kawabata, Kohsuke Imai, Tomohiro Morio, Kazuo Tamura, Akifumi Takaori-Kondo, Masayuki Yamamoto, Satoru Miyano, Seiji Kojima, Etsuro Ito, Seishi Ogawa, Keitaro Matsuo, Hiromasa Yabe, Miharu Yabe, Minoru Takata. Pathogenic mutations identified by a multimodality approach in 117 Japanese Fanconi anemia patients. Haematologica. 2020.04; 105 (4): 1166-1167. ( PubMed, DOI )
- Yao-Zhong Zhang, Arda Akdemir, Georg Tremmel, Seiya Imoto, Satoru Miyano, Tetsuo Shibuya, Rui Yamaguchi. Nanopore basecalling from a perspective of instance segmentation. BMC Bioinformatics. 2020.04; 21 (Suppl 3): 136. ( PubMed, DOI )
- Shunsuke Kimura, Masafumi Seki, Tomoko Kawai, Hiroaki Goto, Kenichi Yoshida, Tomoya Isobe, Masahiro Sekiguchi, Kentaro Watanabe, Yasuo Kubota, Yasuhito Nannya, Hiroo Ueno, Yusuke Shiozawa, Hiromichi Suzuki, Yuichi Shiraishi, Kentaro Ohki, Motohiro Kato, Katsuyoshi Koh, Ryoji Kobayashi, Takao Deguchi, Yoshiko Hashii, Toshihiko Imamura, Atsushi Sato, Nobutaka Kiyokawa, Atsushi Manabe, Masashi Sanada, Marc R Mansour, Akira Ohara, Keizo Horibe, Masao Kobayashi, Akira Oka, Yasuhide Hayashi, Satoru Miyano, Kenichiro Hata, Seishi Ogawa, Junko Takita. DNA methylation-based classification reveals difference between pediatric T-cell acute lymphoblastic leukemia and normal thymocytes. Leukemia. 2020.04; 34 (4): 1163-1168. ( PubMed, DOI )
- Atsushi Niida, Takanori Hasegawa, Hideki Innan, Tatsuhiro Shibata, Koshi Mimori, Satoru Miyano. A unified simulation model for understanding the diversity of cancer evolution. PeerJ. 2020.04; 8 e8842. ( PubMed, DOI )
- Satoru Miyano. Digesting Cancer Big Networks by Explainable AI and Supercomputers. The 3rd R-CCS International Symposium (R-CCSIS'21) 2021.02.16 Online
- 渡邉 健太郎, 加登 翔太, 磯部 知弥, 緒方 瑛人, 田坂 佳資, 上野 浩生, 南谷 泰仁, 田中 洋子, 白石 友一, 千葉 健一, 梅田 雄嗣, 樋渡 光輝, 宮野 悟, 小川 誠司, 滝田 順子. 小児・AYA腫瘍の最近の進歩 難治性骨肉腫における糖鎖遺伝子の役割と治療標的としての可能性. 第79回日本癌学会学術総会 2020.10.03 リーガロイヤルホテル広島
- 渡邉 健太郎、加登 翔太、磯部 知弥、緒方 瑛人、田坂 佳資、上 野 浩生、南谷 泰仁、田中 洋子、白石 友一、千葉 健一、梅田 雄嗣、樋渡 光輝、宮野 悟、小川 誠司、滝田 順子. 難治性骨肉腫における糖鎖遺伝子の役割と治療標的としての可能性. 2020.10.03 リーガロイヤルホテル広島
- 古川 洋一、山口 貴世志、笠島 理加、清水 英悟、高根 希世子、 山口 類、井元 清哉、宮野 悟、池上 恒雄. 消化管遺伝性腫瘍の最前線. 第79回日本癌学会学術総会 2020.10.03
- 宮野 悟. AIとがん研究・医療との対話 - がん研究・医療のための自然言語処理と説明可能 AI. 第79回日本癌学会学術総会 2020.10.02 リーガロイヤルホテル広島
- 垣内 伸之、内野 基、木原 多佳子、赤木 宏太朗、井上 善景、横 山 顕礼、平野 智紀、廣田 誠一、池内 浩基、竹内 理、宮野 悟、妹尾 浩、小川 誠司. 潰瘍性大腸炎における大腸上皮クローン進化から明らかとなった大腸 がんの脆弱性. 第79回日本癌学会学術総会 2020.10.02 リーガロイヤルホテル広島
- 井上 善景, 垣内 伸之, 吉田 健一, 塩澤 裕介, 竹内 康英, 千葉 健一, 吉里 哲一, 長山 聡, 宮野 悟, 坂井 義治, 小川 誠司. POLE遺伝子に変異を持つ大腸癌における免疫回避機構. 日本癌学会総会記事 2020.10.01
- 南谷 泰仁, 牧島 秀樹, 竹田 淳恵, 桃沢 幸秀, 佐伯 龍之介, 宮崎 泰司, 石川 隆之, 大屋敷 一馬, Hellstroem Lindberg Eva, Mario Cazzola, Torsten Haferlach, 鎌谷 洋一郎, 久保 充明, 宮野 悟, 小川 誠司. 骨髄系腫瘍におけるDDX41変異の意義. 日本癌学会総会記事 2020.10.01
- 古川 洋一, 山口 貴世志, 笠島 理加, 清水 英悟, 高根 希世子, 山口 類, 井元 清哉, 宮野 悟, 池上 恒雄. 遺伝性腫瘍-最新情報と今後の方向性 消化管遺伝性腫瘍の最前線. 日本癌学会総会記事 2020.10.01
- 垣内 伸之, 横山 顕礼, 内野 基, 木原 多佳子, 赤木 宏太朗, 井上 善景, 平野 智紀, 廣田 誠一, 池内 浩基, 竹内 理, 宮野 悟, 妹尾 浩, 小川 誠司. 遺伝子変異クローンによる食道および大腸組織の再構築の解明. 第79回日本癌学会学術総会 2020.10.01
- 笠島 理加, 鈴木 理樹, 清水 英悟, 玉田 嘉紀, 新井田 厚司, 山口 類, 井元 清哉, 古川 洋一, 宮野 悟, 横瀬 智之, 宮城 洋平. 胎児性肺癌における遺伝子ネットワーク解析. 日本癌学会総会記事 2020.10.01
- 大津 敬, 笠島 理加, 鷲見 公太, 廣島 幸彦, 片山 琴絵, 清水 英悟, 山口 類, 井元 清哉, 宮野 悟, 河村 大輔, 石川 俊平, 横瀬 智之, 比留間 徹, 宮城 洋平. 肉腫患者由来ゼノグラフトのRNA発現解析. 日本癌学会総会記事 2020.10.01
- 竹内 康英, 鈴木 啓道, 吉田 健一, 白石 友一, 垣内 伸之, 塩澤 裕介, 井上 善景, 千葉 健一, 牧島 秀樹, 宮野 悟, 羽賀 博典, Damm Frederik, 小川 誠司. 粘液線維肉腫にみられるTP53の異常と著明な遺伝的不安定性. 日本癌学会総会記事 2020.10.01
- 鎌谷 高志, 平田 真, 植田 幸嗣, 片山 琴絵, 山口 類, 松原 大祐, 藤田 征志, 高澤 豊, 山下 享子, 中村 卓郎, 井元 清哉, 宮野 悟, 中川 英刀, 松田 浩一, 角田 達彦. 粘液型脂肪肉腫におけるがん微小環境の免疫学的解析. 日本癌学会総会記事 2020.10.01
- 平野 智紀, 垣内 伸之, 竹内 康英, 増井 俊彦, 上本 伸二, 南口 早智子, 羽賀 博典, 白石 友一, 宮野 悟, 宇座 徳光, 児玉 裕三, 妹尾 浩, 千葉 勉, 小川 誠司. 異時性多発膵癌の遺伝子解析. 日本癌学会総会記事 2020.10.01
- 越智 陽太郎, 吉田 健一, 佐々木 光, 細谷 紀子, 塩澤 裕介, 南谷 泰仁, 石川 隆之, 白石 友一, 千葉 健一, 田中 洋子, 真田 昌, 牧島 秀樹, 高折 晃史, 宮野 悟, 三谷 絹子, 小川 誠司. 慢性骨髄性白血病急性転化の遺伝学的機序と予後. 日本癌学会総会記事 2020.10.01
- 竹田 淳恵, 吉田 健一, 依田 成玄, 南谷 泰仁, 中川 正宏, 越智 陽太郎, 昆 彩奈, 吉里 哲一, 塩澤 裕介, 白石 友一, 千葉 健一, 田中 洋子, 真田 昌, 宮野 悟, 牧島 秀樹, 小川 誠司. 急性赤白血病におけるJAK阻害剤の検討. 日本癌学会総会記事 2020.10.01
- 土方 康基, 横山 和明, 山口 貴世志, 池上 恒雄, 山口 類, 井元 清哉, 内丸 薫, 宮野 悟, 四柳 宏, 東條 有伸, 古川 洋一. 当院における標準治療不応進行がん患者を対象としたクリニカルシーケンスの検討. 日本癌学会総会記事 2020.10.01
- 垣内 伸之, 内野 基, 木原 多佳子, 赤木 宏太朗, 井上 善景, 横山 顕礼, 平野 智紀, 廣田 誠一, 池内 浩基, 竹内 理, 宮野 悟, 妹尾 浩, 小川 誠司. 大腸がんの診断・治療と発がん研究における新しい知見 潰瘍性大腸炎における大腸上皮クローン進化から明らかとなった大腸がんの脆弱性. 日本癌学会総会記事 2020.10.01
- 西村 友美, 垣内 伸之, 吉田 健一, 竹内 康英, 前田 紘奈, 塩澤 裕介, 平田 勝啓, 片岡 竜貴, 桜井 孝規, 馬場 郷子, 竹内 賢吾, 羽賀 博典, 宮野 悟, 戸井 雅和, 小川 誠司. 乳がん微小環境の特性と全身性応答、新しい治療展開 乳管上皮増殖性病変から乳癌へ至るクローン進化. 日本癌学会総会記事 2020.10.01
- 藤井 陽一, 佐藤 悠佑, 鈴木 啓道, 吉里 哲一, 垣内 伸之, 吉田 健一, 千葉 健一, 白石 友一, 西松 寛明, 岡根谷 利一, 真田 昌, 南谷 泰仁, 牧島 秀樹, 宮野 悟, 久米 春喜, 小川 誠司. 上部尿路上皮癌の分子分類と新規バイオマーカー. 日本癌学会総会記事 2020.10.01
- 関口 昌央, 関 正史, 吉田 健一, 田中 水緒, 白井 了太, 宗崎 良太, 白石 友一, 樋渡 光輝, 加藤 元博, 田口 智章, 田中 祐吉, 宮野 悟, 小川 誠司, 滝田 順子. マルチオミックス解析による肝芽腫の治療標的NQO1、ODC1の同定. 日本癌学会総会記事 2020.10.01
- 小笠原 辰樹, 藤井 陽一, 塩澤 裕介, 牧島 秀樹, 中村 英二郎, 田中 知明, 白石 友一, 宮野 悟, 小川 誠司. チアノーゼ性先天性心疾患に伴う多発パラガングリオーマは異なるHIF2α変異を伴う平行進化を示した. 日本癌学会総会記事 2020.10.01
- 佐伯 龍之介, 吉里 哲一, 白石 友一, 千葉 健一, 田中 洋子, 松田 浩一, 村上 善則, 久保 充明, 宮野 悟, 牧島 秀樹, 小川 誠司. クローン造血における遺伝子変異とコピー数異常の統合解析. 日本癌学会総会記事 2020.10.01
- 藤田 征志, 山口 類, 有廣 光司, 島田 周, 宮野 悟, 山上 裕機, 茶山 一彰, 垣見 和宏, 田中 真二, 井元 清哉, 中川 英刀. がん免疫ゲノム解析 肝臓がんの免疫ゲノム解析. 日本癌学会総会記事 2020.10.01
- 藤田 征志、山口 類、有廣 光司、島田 周、宮野 悟、山上 裕 機、茶山 一彰、垣見 和宏、田中 真二、井元 清哉2、中川 英刀. 肝臓がんの免疫ゲノム解析. 第79回日本癌学会学術総会 2020.10.01 リーガロイヤルホテル広島
- 前田 紘奈、垣内 伸之、平野 智紀、竹内 康英、伊藤 孝司、 小川 絵里、塩川 雅広、宇座 徳光、田中 洋子、南谷 泰仁、牧 島 秀樹、上本 伸二、宮野 悟、小川 誠司. 慢性炎症に伴う胆管上皮におけるクローン拡大. 第79回日本癌学会学術総会 2020.10.01 リーガロイヤルホテル広島
- Satoru Miyano. Cancer Big Data Challenges from Genomes to Networks. The 24th International Conference on Research in Computational Molecular Biology (RECOBM 2020) 2020.06.23 Online
- Nobuyuki Kakiuchi, Kenichi Yoshida, Motoi Uchino, Takako Kihara, Akaki Kotaro, Yoshikage Inoue, Kenji Kawada, Satoshi Nagayama, Akira Yokoyama, Tomonori Hirano, Yasuhide Takeuchi, Hiroyuki Miyoshi, Yoshiharu Sakai, Hironori Haga, Seiichi Hirota, Hiroki Ikeuchi, Osamu Takeuchi, Satoru Miyano, Hiroshi Seno, Seishi Ogawa. Analysis of clonal expansion in epithelium affected by ulcerative colitis reveals frequent mutations affecting IL-17 signaling pathway and novel cancer vulnerability. AACR Annual Meeting 2020 2020.04.27 Online, Philadelphia, USA
- Hirano Tomonori, Nobuyuki Kakiuchi, Yasuhide Takeuchi, Yoshikage Inoue, Tomomi Nishimura, Yoichi Fujii, Akira Yokoyama, Hideki Makishima, Toshihiko Masui, Shinji Uemoto, Sachiko Minamiguchi, Hironori Haga, Kenichi Chiba, Hiroko Tanaka, Yuichi Shiraishi, Satoru Miyano, Norimitsu Uza, Yuzo Kodama, Hiroshi Seno, Seishi Ogawa. Genetic analysis of metachronous pancreatic cancers. AACR Annual Meeting 2020 2020.04.27 Online, Philadelphia, USA
- Yoichi Fujii, Yusuke Sato, Hiromichi Suzuki, Tetsuichi Yoshizato, Kenichi Yoshida, Yuichi Shiraishi, Taketo Kawai, Tohru Nakagawa, Hiroaki Nishimatsu, Toshikazu Nishimatsu, Masashi Sanada, Hideki Makishima, Satoru Miyano, Haruki Kume, Seishi Ogawa. Distinct molecular subtypes and a high diagnostic urinary biomarker of upper urinary tract urothelial carcinoma. AACR Annual Meeting 2020 2020.04.27 Online, Philadelphia, USA
- Yoshikage Inoue, Nobuyuki Kakiuchi, Kenichi Yoshida, Yasuhide Takeuchi, Yusuke Shiozawa, Yuichi Shiraishi, Kenichi Chiba, Tetsuichi Yoshizato, Hiroko Tanaka, Ai Okada, Satoshi Nagayama, Satoru Miyano, Yoshiharu Sakai, Seishi Ogawa. Frequent genomic alterations to evade the immune system in colorectal cancer with POLE gene mutation. AACR Annual Meeting 2020 2020.04.27 Online, Philadelphia, USA
- Yasuhide Takeuchi, Annegret Kunitz, Adriane Halik, Hiromichi Suzuki, Kenichi Yoshida, Yuichi Shiraishi, Kenichi Chiba, Hiroko Tanaka, Nobuyuki Kakiuchi, Yusuke Shiozawa, Akira Yokoyama, Yoshikage Inoue, Tetsuichi Yoshizato, Kosuke Aoki, Yoichi Fujii, Yasuhito Nannya, Hideki Makishima, Satoru Miyano, Hironori Haga, Frederik Damm, Seishi Ogawa. Frequent abnormalities in TP53 and increased genetic instability in myxofibrosarcoma. AACR Annual Meeting 2020 2020.04.27 Online, Philadelphia, USA
- 第16回ヘルシーソサエティ賞(パイオニア部門),ヘルシー・ソサエティ賞事務局,2020年11月
過去の主な研究業績
※太字が当分野所属
- ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium. Pan-cancer analysis of whole genomes. Nature. 578(7793):82-93, 2020.
- Kakiuchi N, Yoshida K, Uchino M, Kihara T, Akaki K, Inoue Y, Kawada K, Nagayama S, Yokoyama A, Yamamoto S, Matsuura M, Horimatsu T, Hirano T, Goto N, Takeuchi Y, Ochi Y, Shiozawa Y, Kogure Y, Watatani Y, Fujii Y, Kim SK, Kon A, Kataoka K, Yoshizato T, MM Nakagawa, Yoda A, Nannya Y, Makishima H, Shiraishi Y, Chiba K, Tanaka H, Sanada M, Sugihara E, Sato T, Maruyama T, Miyoshi H, Taketo MM, Oishi J, Inagaki R, Ueda Y, Okamoto S, Okajima H, Sakai Y, Sakurai T, Haga H, Hirota S, Ikeuchi H, Nakase H, Marusawa H, Chiba T, Takeuchi O, Miyano S, Seno H, Ogawa S. Frequent mutations converging into NFKBIZ signalling in ulcerative colitis. Nature. 577:260–265, 2020.
- Ochi Y, Kon A, Sakata T, Nakagawa MM, Nakazawa N, Kakuta M, Kataoka K, Koseki H, Nakayama M, Morishita D, Tsuruyama T, Saiki R, Yoda A, Okuda R, Yoshizato T, Yoshida K, Shiozawa Y, Nannya Y, Kotani S, Kogure Y, Kakiuchi N, Nishimura T, Makishima H, Malcovati L, Yokoyama A, Takeuchi K, Sugihara E, Sato TA, Sanada M, Takaori-Kondo A, Cazzola M, Kengaku M, Miyano S, Shirahige K, Suzuki HI, Ogawa S. Combined cohesin-Runx1 deficiency synergistically perturbs chromatin looping and causes myelodysplastic syndromes. Cancer Discov. pii: CD-19-0982. doi: 10.1158/2159-8290, 2020.
- Yokoyama A, Kakiuchi N, Yoshizato T, Nannya Y, Suzuki H, Takeuchi Y, Shiozawa Y, Sato Y, Aoki K, Kim SK, Fujii Y, Yoshida K, Kataoka K, Nakagawa MM, Inoue Y, Hirano T, Shiraishi Y, Chiba K, Tanaka H, Sanada M, Nishikawa Y, Amanuma Y, Ohashi S, Aoyama I, Horimatsu T, Miyamoto S, Tsunoda S, Sakai Y, Narahara M, Brown JB, Sato Y, Sawada G, Mimori K, Minamiguchi S, Haga H, Seno H, Miyano S, Makishima H, Muto M, Ogawa S. Age-related remodelling of oesophageal epithelia by mutated cancer drivers. Nature. 565(7739):312-317, 2019.
- Okuno Y, Murata T, Sato Y, Muramatsu H, Ito Y, Watanabe T, Okuno T, Murakami N, Yoshida K, Sawada A, Inoue M, Kawa K, Seto M, Ohshima K, Shiraishi Y, Chiba K, Tanaka H, Miyano S, Narita Y, Yoshida M, Goshima F, Kawada JI, Nishida T, Kiyoi H, Kato S, Nakamura S, Morishima S, Yoshikawa T, Fujiwara S, Shimizu N, Isobe Y, Noguchi M, Kikuta A, Iwatsuki K, Takahashi Y, Kojima S, Ogawa S, Kimura H. Defective Epstein-Barr virus in chronic active infection and haematological malignancy. Nature Microbiology. 4(3):404-413, 2019; Publisher Correction: Nature Microbiology. 2019 Mar;4(3):544.
- Matsuno Y, Atsumi Y, Shimizu A, Katayama K, Fujimori H, Hyodo M, Minakawa Y, Nakatsu Y, Kaneko S, Hamamoto R, Shimamura T, Miyano S, Tsuzuki T, Hanaoka F, Yoshioka KI. Replication stress triggers microsatellite destabilization and hypermutation leading to clonal expansion in vitro. Nature Commun. 10(1):3925, 2019.
- Kajino T, Shimamura T, Gong S, Yanagisawa K, Ida L, Nakatochi M, Griesing S, Shimada Y, Kano K, Suzuki M, Miyano S, Takahashi T. Divergent lncRNA MYMLR regulates MYC by eliciting DNA looping and promoter-enhancer interaction. EMBO J. 8(17):e98441, 2019.
- Saito T, Niida A, Uchi R, Hirata H, Komatsu H, Sakimura S, Hayashi S, Nambara S, Kuroda Y, Ito S, Eguchi H, Masuda T, Sugimachi K, Tobo T, Nishida H, Daa T, Chiba K, Shiraishi Y, Yoshizato T, Kodama M, Okimoto T, Mizukami K, Ogawa R, Okamoto K, Shuto M, Fukuda K, Matsui Y, Shimamura T, Hasegawa T, Doki Y, Nagayama S, Yamada K, Kato M, Shibata T, Mori M, Aburatani H, Murakami K, Suzuki Y, Ogawa S, Miyano S, Mimori K. A temporal shift of the evolutionary principle shaping intratumor heterogeneity in colorectal cancer. Nature Commun. 9(1):2884, 2018.
- Shiozawa Y, Malcovati L, Gallì A, Sato-Otsubo A, Kataoka K, Sato Y, Watatani Y, Suzuki H, Yoshizato T, Yoshida K, Sanada M, Makishima H, Shiraishi Y, Chiba K, Hellström-Lindberg E, Miyano S, Ogawa S, Cazzola M. Aberrant splicing and defective mRNA production induced by somatic spliceosome mutations in myelodysplasia. Nature Commun. 9(1):3649, 2018.
- da Silva-Coelho P, Kroeze LI, Yoshida K, Koorenhof-Scheele TN, Knops R, van de Locht LT, de Graaf AO, Massop M, Sandmann S, Dugas M, Stevens-Kroef MJ, Cermak J, Shiraishi Y, Chiba K, Tanaka H, Miyano S, de Witte T, Blijlevens NMA, Muus P, Huls G, van der Reijden BA, Ogawa S, Jansen JH. Clonal evolution in myelodysplastic syndromes. Nature Commun. 8:15099, 2017.
- Makishima H, Yoshizato T, Yoshida K, Sekeres MA, Radivoyevitch T, Suzuki H, Przychodzen B, Nagata Y, Meggendorfer M, Sanada M, Okuno Y, Hirsch C, Kuzmanovic T, Sato Y, Sato-Otsubo A, LaFramboise T, Hosono N, Shiraishi Y, Chiba K, Haferlach C, Kern W, Tanaka H, Shiozawa Y, Gómez-Seguí I, Husseinzadeh HD, Thota S, Guinta KM, Dienes B, Nakamaki T, Miyawaki S, Saunthararajah Y, Chiba S, Miyano S, Shih LY, Haferlach T, Ogawa S, Maciejewski JP. Dynamics of clonal evolution in myelodysplastic syndromes. Nature Genetics. 49(2):204-212, 2017.
- Seki M, Kimura S, Isobe T, Yoshida K, Ueno H, Nakajima-Takagi Y, Wang C, Lin L, Kon A, Suzuki H, Shiozawa Y, Kataoka K, Fujii Y, Shiraishi Y, Chiba K, Tanaka H, Shimamura T, Masuda K, Kawamoto H, Ohki K, Kato M, Arakawa Y, Koh K, Hanada R, Moritake H, Akiyama M, Kobayashi R, Deguchi T, Hashii Y, Imamura T, Sato A, Kiyokawa N, Oka A, Hayashi Y, Takagi M, Manabe A, Ohara A, Horibe K, Sanada M, Iwama A, Mano H, Miyano S, Ogawa S, Takita J. Recurrent SPI1 (PU.1) fusions in high-risk pediatric T cell acute lymphoblastic leukemia. Nature Genetics. 49(8):1274-1281, 2017.
- Fujimoto A, Furuta M, Totoki Y, Tsunoda T, Kato M, Shiraishi Y, Tanaka H, Taniguchi H, Kawakami Y, Ueno M, Gotoh K, Ariizumi S, Wardell CP, Hayami S, Nakamura T, Aikata H, Arihiro K, Boroevich KA, Abe T, Nakano K, Maejima K, Sasaki-Oku A, Ohsawa A, Shibuya T, Nakamura H, Hama N, Hosoda F, Arai Y, Ohashi S, Urushidate T, Nagae G, Yamamoto S, Ueda H, Tatsuno K, Ojima H, Hiraoka N, Okusaka T, Kubo M, Marubashi S, Yamada T, Hirano S, Yamamoto M, Ohdan H, Shimada K, Ishikawa O, Yamaue H, Chayama K, Miyano S, Aburatani H, Shibata T, Nakagawa H. Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nature Genetics. 48(5):500-509, 2016. Erratum: Nature Genetics. 48(6):700. doi: 10.1038/ng0616-700a.
- Kataoka K, Shiraishi Y, Takeda Y, Sakata S, Matsumoto M, Nagano S, Maeda T, Nagata Y, Kitanaka A, Mizuno S, Tanaka H, Chiba K, Ito S, Watatani Y, Kakiuchi N, Suzuki H, Yoshizato T, Yoshida K, Sanada M, Itonaga H, Imaizumi Y, Totoki Y, Munakata W, Nakamura H, Hama N, Shide K, Kubuki Y, Hidaka T, Kameda T, Masuda K, Minato N, Kashiwase K, Izutsu K, Takaori-Kondo A, Miyazaki Y, Takahashi S, Shibata T, Kawamoto H, Akatsuka Y, Shimoda K, Takeuchi K, Seya T, Miyano S, Ogawa S. Aberrant PD-L1 expression through 3'-UTR disruption in multiple cancers. Nature. 534(7607):402-406, 2016.
- Fujimoto A, Furuta M, Shiraishi Y, Gotoh K, Kawakami Y, Arihiro K, Nakamura T, Ueno M, Ariizumi SI, Hai Nguyen H, Shigemizu D, Abe T, Boroevich KA, Nakano K, Sasaki A, Kitada R, Maejima K, Yamamoto Y, Tanaka H, Shibuya T, Shibata T, Ojima H, Shimada K, Hayami S, Shigekawa Y, Aikata H, Ohdan H, Marubashi S, Yamada T, Kubo M, Hirano S, Ishikawa O, Yamamoto M, Yamaue H, Chayama K, Miyano S, Tsunoda T, Nakagawa H. Whole-genome mutational landscape of liver cancers displaying biliary phenotype reveals hepatitis impact and molecular diversity. Nature Commun. 6:6120, 2015.
- Kataoka K, Nagata Y, Kitanaka A, Shiraishi Y, Shimamura T, Yasunaga J, Totoki Y, Chiba K, Sato-Otsubo A, Nagae G, Ishii R, Muto S, Kotani S, Watatani Y, Takeda J, Sanada M, Tanaka H, Suzuki H, Sato Y, Shiozawa Y, Yoshizato T, Yoshida K, Makishima H, Iwanaga M, Ma G, Nosaka K, Hishizawa M, Itonaga H, Imaizumi Y, Munakata W, Ogasawara H, Sato T, Sasai K, Muramoto K, Penova M, Kawaguchi T, Nakamura H, Hama N, Shide K, Kubuki Y, Hidaka T, Kameda T, Nakamaki T, Ishiyama K, Miyawaki S, Yoon SS, Tobinai K, Miyazaki Y, Takaori-Kondo A, Matsuda F, Takeuchi K, Nureki O, Aburatani H, Watanabe T, Shibata T, Matsuoka M, Miyano S, Shimoda K, Ogawa S. Integrated molecular analysis of adult T cell leukemia/lymphoma. Nature Genetics. 47(11):1304-1315, 2015.
- Madan V, Kanojia D, Li J, Okamoto R, Sato-Otsubo A, Kohlmann A, Sanada M, Grossmann V, Sundaresan J, Shiraishi Y, Miyano S, Thol F, Ganser A, Yang H, Haferlach T, Ogawa S, Koeffler HP. Aberrant splicing of U12-type introns is the hallmark of ZRSR2 mutant myelodysplastic syndrome. Nature Commun. 6:6042, 2015.
- Polprasert C, Schulze I, Sekeres MA, Makishima H, Przychodzen B, Hosono N, Singh J, Padgett RA, Gu X, Phillips JG, Clemente M, Parker Y, Lindner D, Dienes B, Jankowsky E, Saunthararajah Y, Du Y, Oakley K, Nguyen N, Mukherjee S, Pabst C, Godley LA, Churpek JE, Pollyea DA, Krug U, Berdel WE, Klein HU, Dugas M, Shiraishi Y, Chiba K, Tanaka H, Miyano S, Yoshida K, Ogawa S, Müller-Tidow C, Maciejewski JP. Inherited and somatic defects in DDX41 in myeloid neoplasms. Cancer Cell. 27(5):658-670, 2015.
- Seki M, Nishimura R, Yoshida K, Shimamura T, Shiraishi Y, Sato Y, Kato M, Chiba K, Tanaka H, Hoshino N, Nagae G, Shiozawa Y, Okuno Y, Hosoi H, Tanaka Y, Okita H, Miyachi M, Souzaki R, Taguchi T, Koh K, Hanada R, Kato K, Nomura Y, Akiyama M, Oka A, Igarashi T, Miyano S, Aburatani H, Hayashi Y, Ogawa S, Takita J. Integrated genetic and epigenetic analysis defines novel molecular subgroups in rhabdomyosarcoma. Nature Commun. 6:7557, 2015.
- Suzuki H, Aoki K, Chiba K, Sato Y, Shiozawa Y, Shiraishi Y, Shimamura T, Niida A, Motomura K, Ohka F, Yamamoto T, Tanahashi K, Ranjit M, Wakabayashi T, Yoshizato T, Kataoka K, Yoshida K, Nagata Y, Sato-Otsubo A, Tanaka H, Sanada M, Kondo Y, Nakamura H, Mizoguchi M, Abe T, Muragaki Y, Watanabe R, Ito I, Miyano S, Natsume A, Ogawa S. Mutational landscape and clonal architecture in grade II and III gliomas. Nature Genetics. 47(5):458-468, 2015.
- Yoshizato T, Dumitriu B, Hosokawa K, Makishima H, Yoshida K, Townsley D, Sato-Otsubo A, Sato Y, Liu D, Suzuki H, Wu CO, Shiraishi Y, Clemente MJ, Kataoka K, Shiozawa Y, Okuno Y, Chiba K, Tanaka H, Nagata Y, Katagiri T, Kon A, Sanada M, Scheinberg P, Miyano S, Maciejewski JP, Nakao S, Young NS, Ogawa S. Somatic mutations and clonal hematopoiesis in aplastic anemia. N Engl J Med. 373(1):35-47, 2015.
- Arima C, Kajino T, Tamada Y, Imoto S, Shimada Y, Nakatochi M, Suzuki M, Isomura H, Yatabe Y, Yamaguchi T, Yanagisawa K, Miyano S, Takahashi T. Lung adenocarcinoma subtypes definable by lung development-related miRNA expression profiles in association with clinicopathologic features. Carcinogenesis. 35(10): 2224-2231, 2014.
- Sakata-Yanagimoto M, Enami T, Yoshida K, Shiraishi Y, Ishii R, Miyake Y, Muto H, Tsuyama N, Sato-Otsubo A, Okuno Y, Sakata S, Kamada Y, Nakamoto-Matsubara R, Tran NB, Izutsu K, Sato Y, Ohta Y, Furuta J, Shimizu S, Komeno T, Sato Y, Ito T, Noguchi M, Noguchi E, Sanada M, Chiba K, Tanaka H, Suzukawa K, Nanmoku T, Hasegawa Y, Nureki O, Miyano S, Nakamura N, Takeuchi K, Ogawa S, Chiba S. Somatic RHOA mutation in angioimmunoblastic T cell lymphoma. Nature Genetics. 46(2):171-175, 2014.
- Sato Y, Maekawa S, Ishii R, Sanada M, Morikawa T, Shiraishi Y, Yoshida K, Nagata Y, Sato-Otsubo A, Yoshizato T, Suzuki H, Shiozawa Y, Kataoka K, Kon A, Aoki K, Chiba K, Tanaka H, Kume H, Miyano S, Fukayama M, Nureki O, Homma Y, Ogawa S. Recurrent somatic mutations underlie corticotropin-independent Cushing's syndrome. Science. 344(6186):917-920, 2014.
- Kon A, Shih LY, Minamino M, Sanada M, Shiraishi Y, Nagata Y, Yoshida K, Okuno Y, Bando M, Nakato R, Ishikawa S, Sato-Otsubo A, Nagae G, Nishimoto A, Haferlach C, Nowak D, Sato Y, Alpermann T, Nagasaki M, Shimamura T, Tanaka H, Chiba K, Yamamoto R, Yamaguchi T, Otsu M, Obara N, Sakata-Yanagimoto M, Nakamaki T, Ishiyama K, Nolte F, Hofmann WK, Miyawaki S, Chiba S, Mori H, Nakauchi H, Koeffler HP, Aburatani H, Haferlach T, Shirahige K, Miyano S, Ogawa S. Recurrent mutations in multiple components of the cohesin complex in myeloid neoplasms. Nature Genetics. 45(10):1232-1237, 2013.
- Makishima H, Yoshida K, Nguyen N, Przychodzen B, Sanada M, Okuno Y, Ng KP, Gudmundsson KO, Vishwakarma BA, Jerez A, Gomez-Segui I, Takahashi M, Shiraishi Y, Nagata Y, Guinta K, Mori H, Sekeres MA, Chiba K, Tanaka H, Muramatsu H, Sakaguchi H, Paquette RL, McDevitt MA, Kojima S, Saunthararajah Y, Miyano S, Shih LY, Du Y, Ogawa S, Maciejewski JP. Somatic SETBP1 mutations in myeloid malignancies. Nature Genetics. 45(8):942-946, 2013.
- Sakaguchi H, Okuno Y, Muramatsu H, Yoshida K, Shiraishi Y, Takahashi M, Kon A, Sanada M, Chiba K, Tanaka H, Makishima H, Wang X, Xu Y, Doisaki S, Hama A, Nakanishi K, Takahashi Y, Yoshida N, Maciejewski JP, Miyano S, Ogawa S, Kojima S. Exome sequencing identifies secondary mutations of SETBP1 and JAK3 in juvenile myelomonocytic leukemia. Nature Genetics. 45(8):937-941, 2013.
- Sato Y, Yoshizato T, Shiraishi Y, Maekawa S, Okuno Y, Kamura T, Shimamura T, Sato-Otsubo A, Nagae G, Suzuki H, Nagata Y, Yoshida K, Kon A, Suzuki Y, Chiba K, Tanaka H, Niida A, Fujimoto A, Tsunoda T, Morikawa T, Maeda D, Kume H, Sugano S, Fukayama M, Aburatani H, Sanada M, Miyano S, Homma Y, Ogawa S. Integrated molecular analysis of clear-cell renal cell carcinoma. Nature Genetics. 45(8):860-867, 2013.
- Yoshida K, Toki T, Okuno Y, Kanezaki R, Shiraishi Y, Sato-Otsubo A, Sanada M, Park MJ, Terui K, Suzuki H, Kon A, Nagata Y, Sato Y, Wang R, Shiba N, Chiba K, Tanaka H, Hama A, Muramatsu H, Hasegawa D, Nakamura K, Kanegane H, Tsukamoto K, Adachi S, Kawakami K, Kato K, Nishimura R, Izraeli S, Hayashi Y, Miyano S, Kojima S, Ito E, Ogawa S. The landscape of somatic mutations in Down syndrome-related myeloid disorders. Nature Genetics. 45:1293–1299, 2013. Erratum in: Nature Genetics. 45(12):1516, 2013.
- Yoshimaru T, Komatsu M, Matsuo T, Chen YA, Murakami Y, Mizuguchi K, Mizohata E, Inoue T, Akiyama M, Yamaguchi R, Imoto S, Miyano S, Miyoshi Y, Sasa M, Nakamura Y, Katagiri T. Targeting BIG3-PHB2 interaction to overcome tamoxifen resistance in breast cancer cells. Nature Commun. 4:2443, 2013.
- Fujimoto A, Totoki Y, Abe T, Boroevich KA, Hosoda F, Hai Nguyen H, Aoki M, Hosono N , Kubo M, Miya F, Arai Y, Takahashi H, Shirakihara T, Nagasaki M, Shibuya T, Nakano K, Watanabe-Makino K, Tanaka H, Nakamura H, Kusuda J, Ojima H , Shimada K, Okusaka T, Ueno M, Shigekawa Y, Kawakami Y, Arihiro K, Ohdan H, Gotoh K, Ishikawa O, Ariizumi S, Yamamoto M, Yamada T, Chayama K, Kosuge T, Yamaue H, Kamatani N, Miyano S, Nakagama H, Nakamura Y, Tsunoda T, Shibata T, Nakagawa H. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nature Genetics . 44(7):760-764, 2012 .
- Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, Sato Y, Sato-Otsubo A, Kon A, Nagasaki M, Chalkidis G, Suzuki Y, Shiosaka M, Kawahata R, Yamaguchi T, Otsu M, Obara N, Sakata-Yanagimoto M, Ishiyama K, Mori H, Nolte F, Hofmann WK, Miyawaki S, Sugano S, Haferlach C, Koeffler HP, Shih LY, Haferlach T, Chiba S, Nakauchi H, Miyano S, Ogawa S. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 478(7367): 64-69, 2011.
- Fujimoto A, Nakagawa H, Hosono N, Nakano K, Abe G, Boroevich KA, Nagasaki M, Yamaguchi R, Shibuya T, Kubo M, Miyano S, Nakamura Y, Tsunoda T. Whole-genome sequencing and comprehensive variant analysis of a Japanese individual using massively parallel sequencing. Nature Genetics. 42: 931–936, 2010.
- Fujita M, Yamaguchi R, Hasegawa T, Shimada S, Arihiro K, Hayashi S, Maejima K, Nakano K, Fujimoto A, Ono A, Aikata H, Ueno M, Hayami S, Tanaka H, Miyano S, Yamaue H, Chayama K, Kakimi K, Tanaka S, Imoto S, Nakagawa H. Classification of primary liver cancer with immunosuppression mechanisms and correlation with genomic alterations. EBioMedicine. 2020 Mar;53:102659. doi: 10.1016/j.ebiom.2020.102659.
- Fujimoto A, Fujita M, Hasegawa T, Wong JH, Maejima K, Oku-Sasaki A, Nakano K, Shiraishi Y, Miyano S, Yamamoto G, Akagi K, Imoto S, Nakagawa H. Comprehensive analysis of indels in whole-genome microsatellite regions and microsatellite instability across 21 cancer types. Genome Res. 2020 Mar 24. doi: 10.1101/gr.255026.119.
- Hasegawa T, Yamaguchi R, Kakuta M, Sawada K, Kawatani K, Murashita K, Nakaji S, Imoto S. Prediction of blood test values under different lifestyle scenarios using time-series electronic health record. PLoS One. 15(3):e0230172, 2020.
- Sato N, Kakuta M, Uchino E, Hasegawa T, Kojima R, Kobayashi W, Sawada K, Tamura Y, Tokuda I, Imoto S, Nakaji S, Murashita K, Yanagita M, Okuno Y. The relationship between cigarette smoking and the tongue microbiome in an East Asian population. J Oral Microbiol. 12(1):1742527, 2020.
- Sato N, Kakuta M, Hasegawa T, Yamaguchi R, Uchino E, Kobayashi W, Sawada K, Tamura Y, Tokuda I, Murashita K, Nakaji S, Imoto S, Yanagita M, Okuno Y. Metagenomic analysis of bacterial species in tongue microbiome of current and never smokers. NPJ Biofilms Microbiomes. 6(1):11, 2020.
- Misawa K, Hasegawa T, Mishima E, Jutabha P, Ouchi M, Kojima K, Kawai Y, Matsuo M, Anzai N, Nagasaki M. Contribution of rare variants of the SLC22A12 gene to the missing heritability of serum urate levels. Genetics. 214(4):1079-1090, 2020.
- Ito S, Yadome M, Nishiki T, Ishiduka S, Yamaguchi R, Miyano S. Virtual Grid Engine: A simulated grid engine environment for large-scale supercomputers. BMC Bioinformatics. 20(Suppl 16):591, 2019.
- Moriyama T, Imoto S, Hayashi S, Shiraishi Y, Miyano S, Yamaguchi R. A Bayesian model integration for mutation calling through data partitioning. Bioinformatics. 35(21):4247-4254, 2019.
- Hayashi S, Moriyama T, Yamaguchi R, Mizuno S, Komura M, Miyano S, Nakagawa H, Imoto S. ALPHLARD-NT: Bayesian method for human leukocyte antigen genotyping and mutation calling through simultaneous analysis of normal and tumor whole-genome sequence data. J Comput Biol. 26(9):923-937, 2019.
- Hayashi S, Yamaguchi R, Mizuno S, Komura M, Miyano S, Nakagawa H, Imoto S. ALPHLARD: a Bayesian method for analyzing HLA genes from whole genome sequence data. BMC Genomics. 19(1):790, 2018.
- Shiraishi Y, Kataoka K, Chiba K, Okada A, Kogure Y, Tanaka H, Ogawa S, Miyano S. A comprehensive characterization of cis-acting splicing-associated variants in human cancer. Genome Res. 28(8):1111-1125, 2018.
- Moriyama T, Shiraishi Y, Chiba K, Yamaguchi R, Imoto S, Miyano S. OVarCall: Bayesian mutation calling method utilizing overlapping paired-end reads. IEEE Trans Nanobioscience. 16(2): 116-122, 2017.
- Zhang YZ, Yamaguchi R, Imoto S, Miyano S. Sequence-specific bias correction for RNA-seq data using recurrent neural networks. BMC Genomics. 18(Suppl 1):1044, 2017.
- Zhang YZ, Imoto S, Miyano S, Yamaguchi R. Reconstruction of high read-depth signals from low-depth whole genome sequencing data using deep learning. BIBM 2017: 1227-1232, 2017.
- Hasegawa T, Kojima K, Kawai Y, Misawa K, Mimori T, Nagasaki M. AP-SKAT: highly-efficient genome-wide rare variant association test. BMC Genomics. 17(1):745, 2016.
- Kojima K, Kawai Y, Nariai N, Mimori T, Hasegawa T, Nagasaki M. Short tandem repeat number estimation from paired-end reads for multiple individuals by considering coalescent tree. BMC Genomics. 17 Suppl 5:494, 2016.
- Koshiba S, Motoike I, Kojima K, Hasegawa T, Shirota M, Saito T, Saigusa D, Danjoh I, Katsuoka F, Ogishima S, Kawai Y, Yamaguchi-Kabata Y, Sakurai M, Hirano S, Nakata J, Motohashi H, Hozawa A, Kuriyama S, Minegishi N, Nagasaki M, Takai-Igarashi T, Fuse N, Kiyomoto H, Sugawara J, Suzuki Y, Kure S, Yaegashi N, Tanabe O, Kinoshita K, Yasuda J, Yamamoto M. The structural origin of metabolic quantitative diversity. Sci Rep. 6:31463, 2016.
- Chiba K, Shiraishi Y, Nagata Y, Yoshida K, Imoto S, Ogawa S, Miyano S. Genomon ITDetector: A tool for somatic internal tandem duplication detection from cancer genome sequencing data. Bioinformatics. 31(1), 116-118, 2015.
- Ito S, Shiraishi Y, Shimamura T, Chiba K, Miyano S. High performance computing of a fusion gene detection pipeline on the K computer. BIBM 2015: 1441-1447, 2015.
- Shiraishi Y, Sato Y, Chiba K, Okuno Y, Nagata Y, Yoshida K, Shiba N, Hayashi Y, Kume H, Homma Y, Sanada M, Ogawa S, Miyano S. An empirical Bayesian framework for somatic mutation detection from cancer. Nucleic Acids Res. 41(7): e89, 2013.
- Niida A, Imoto S, Shimamura T, Miyano S. Statistical model-based testing to evaluate the recurrence of genomic aberrations. Bioinformatics. 28(12):i115-i120, 2012.
- Shiraishi U, Okada-Hatakeyama M, Miyano S. A rank-based statistical test for measuring synergistic effects between two gene sets. Bioinformatics. 27 (17): 2399-2405, 2011.
- Hasegawa T, Yamaguchi R, Niida A, Miyano S, Imoto S. Ensemble smoothers for inference of hidden states and parameters in combinatorial regulatory model. J Franklin Institute. (5):2916-2933, 2020.
- Niida A, Hasegawa T, Innan H, Shibata T, Mimori K, Miyano S. A unified simulation model for understanding the diversity of cancer evolution. PeerJ. 8:e8842, 2020.
- Niida A, Hasegawa T, Miyano S. Sensitivity analysis of agent-based simulation utilizing massively parallel computation and interactive data visualization. PLoS One. 14(3):e0210678, 2019.
- Park H, Yamada M, Imoto S, Miyano S. Robust sample-specific stability selection with effective error control. J Comput Biol. 26(3):202-217, 2019.
- Park H, Shimamura T, Imoto S, Miyano S. Adaptive NetworkProfiler for identifying cancer characteristic-specific gene regulatory networks. J Comput Biol. 25(2):130-145, 2018.
- Mimori K, Saito T, Niida A, Miyano S. Cancer evolution and heterogeneity. Ann Gastroenterol Surg. 2(5):332-338, 2018.
- Hasegawa T, Kojima K, Kawai Y, Nagasaki M. Time-series filtering for replicated observations via a kernel approximate Bayesian computation. IEEE Transactions on Signal Processing. 66(23):6148-6161, 2018.
- Niida A, Nagayama S, Miyano S, Mimori K. Understanding intratumor heterogeneity by combining genome analysis and mathematical modeling. Cancer Sci. 109(4):884-892, 2018.
- Park H, Niida A, Imoto S, Miyano S. Interaction-based feature selection for uncovering cancer driver genes through copy number-driven expression level. J Comput Biol. 24(2):138-152, 2017.
- Park H, Shiraishi Y, Imoto S, Miyano S. A novel adaptive penalized logistic regression for uncovering biomarker associated with anti-cancer drug sensitivity. IEEE/ACM Trans Comput Biol Bioinform. 14(4):771-782, 2017.
- Matsui Y, Niida A, Uchi R, Mimori K, Miyano S, Shimamura T. phyC: Clustering cancer evolutionary trees. PLoS Comput Biol. 13(5):e1005509, 2017.
- Morita T, Rahman A, Hasegawa T, Ozaki A, Tanimoto T. The potential possibility of symptom checker. Int J Health Policy Manag. 6(10):615-616, 2017.
- Kurashige J, Hasegawa T, Niida A, Sugimachi K, Deng N, Mima K, Uchi R, Sawada G, Takahashi Y, Eguchi H, Inomata M, Kitano S, Fukagawa T, Sasako M, Sasaki H, Sasaki S, Mori M, Yanagihara K, Baba H, Miyano S, Tan P, Mimori K. Integrated molecular profiling of human gastric cancer identifies DDR2 as a potential regulator of peritoneal dissemination. Sci Rep. 6:22371, 2016.
- Hasegawa T, Mori T, Yamaguchi R, Shimamura T, Miyano S, Imoto S, Akutsu T. Genomic data assimilation using a higher moment filtering technique for restoration of gene regulatory networks. BMC Syst Biol. 9:14, 2015.
- Park H, Niida, A, Miyano S, Imoto S. Sparse overlapping group lasso for integrative multi-omics analysis. J Comput Biol. 22(2): 73-84, 2015.
- Park H, Imoto S, Miyano S. Recursive Random Lasso (RRLasso) for identifying anti-cancer drug targets. PLoS One. 10(11):e0141869, 2015.
- Nakata A, Yoshida R, Yamaguchi R, Yamauchi M, Tamada Y, Fujita A, Shimamura T, Imoto S, Higuchi T, Nomura M, Kimura T, Nokihara H, Higashiyama M, Kondoh K, Nishihara H, Tojo A, Yano S, Miyano S, Gotoh N. Elevated β-catenin pathway as a novel target for patients with resistance to EGF receptor targeting drugs. Sci Rep. 5:13076, 2015.
- Hasegawa T, Yamaguchi R, Nagasaki M, Miyano S, Imoto S. Inference of gene regulatory networks incorporating multi-source biological knowledge via a state space model with L1 regularization. PLoS One. 9(8):e105942, 2014.
- Hasegawa T, Nagasaki M, Yamaguchi R, Imoto S, Miyano S. An efficient method of exploring simulation models by assimilating literature and biological observational data. Biosystems. 121:54-66, 2014.
- Hasegawa T, Mori T, Yamaguchi R, Imoto S, Miyano S, Akutsu T. An efficient data assimilation schema for restoration and extension of gene regulatory networks using time-course observation data. J Comput Biol. 21(11):785-798, 2014.
- Park H, Shimamura T, Miyano S, Imoto S. Robust prediction of anti-cancer drug sensitivity and sensitivity-specific biomarker. PLoS One. 9(1): e108990, 2014.
- Yamauchi M, Yamaguchi R, Nakata A, Kohno T, Nagasaki M, Shimamura T, Imoto S, Saito A, Ueno K, Hatanaka Y, Yoshida R, Higuchi T, Nomura M, Beer DG, Yokota J, Miyano S, Gotoh N. Epidermal growth factor receptor tyrosine kinase defines critical prognostic genes of stage I lung adenocarcinoma. PLoS One. 7(9): e43923, 2012.
- Shimamura T, Imoto S, Shimada Y, Hosono Y, Niida A, Nagasaki M, Yamaguchi R, Takahashi T, Miyano S. A novel network profiling analysis reveals system changes in epithelial-mesenchymal transition. PLoS One. 6(6): e20804, 2011.
- Tamada Y, Imoto S, Miyano S. Parallel algorithm for learning optimal Bayesian network structure. J Machine Learning Research. 12: 2437-2459, 2011.
- Tamada Y, Imoto S, Araki H, Nagasaki M, Print C, Charnock-Jones DS, Miyano S. Estimating genome-wide gene networks using nonparametric Bayesian network models on massively parallel computers. IEEE/ACM Transactions on Computational Biology and Bioinformatics. 8(3): 683 - 697, 2011. Tamada Y, Shimamura T, Yamaguchi R, Imoto S, Nagasaki M, Miyano S. SiGN: large-scale gene network estimation environment for high performance computing. Genome Informatics. 25: 40-52, 2011.
- Tamada Y, Yamaguchi R, Imoto S, Hirose O, Yoshida R, Nagasaki M, Miyano S. SiGN-SSM: open source parallel software for estimating gene networks with state space models. Bioinformatics. 27: 1172-1173, 2011.
- Kojima K, Perrier E, Imoto S, Miyano S. Optimal search on clustered structural constraint for learning Bayesian network structure. J Machine Learning Research. 11: 285−310, 2010.
- Yamaguchi R, Imoto S, Yamauchi M, Nagasaki M, Yoshida R, Shimamura T, Hatanaka Y, Ueno K, Higuchi T, Gotoh N, Miyano S. Predicting differences in gene regulatory systems by state space models. Genome Informatics. 21:101-113, 2008.
- Perrier E, Imoto S, Miyano S. Finding optimal Bayesian network given a super-structure. J Machine Learning Research. 9: 2251-2286, 2008.
- Yamaguchi R, Yoshida R, Imoto S, Higuchi T, Miyano S. Finding module-based gene networks with state-space models? Mining high-dimensional and short time-course gene expression data. IEEE Signal Processing Magazine. 24(1): 37-46, 2007.
- Imoto S, Tamada Y, Araki H, Yasuda K, Print CG, Charnock-Jones SD, Sanders D, Savoie CJ, Tashiro K, Kuhara S, Miyano S. Computational strategy for discovering druggable gene networks from genome-wide RNA expression profiles. Pacific Symposium on Biocomputing. 11: 559-571, 2006.
- Ott S, Hansen A, Kim S-Y, Miyano S. Superiority of network motifs over optimal networks and an application to the revelation of gene network evolution. Bioinformatics. 21(2): 227-238, 2005.
- Imoto S, Goto T, Miyano S. Estimation of genetic networks and functional structures between genes by using Bayesian network and nonparametric regression. Pacific Symposium on Biocomputing. 7:175-186, 2002.
- Anderson WP; Global Life Science Data Resources Working Group: Apweiler R, Bateman A, Bauer GA, Berman H, Blake JA, Blomberg N, Burley SK, Cochrane G, Di Francesco V, Donohue T, Durinx C, Game A, Green E, Gojobori T, Goodhand P, Hamosh A, Hermjakob H, Kanehisa M, Kiley R, McEntyre J, McKibbin R, Miyano S, Pauly B, Perrimon N, Ragan MA, Richards G, Teo YY, Westerfield M, Westhof E, Lasko PF. Data management: A global coalition to sustain core data. Nature. 543(7644):179, 2017.
AI技術開発分野

生物統計学分野

データ科学アルゴリズム
設計・解析分野
