Department of AI Systems Medicine
Member

Hideyuki SHIMIZU
Professor
Satoshi Ohno
Junior Associate Professor
Hidekazu Hishinuma
Assistant Professor

Research Projects
AI Drug Discovery
We developed LIGHTHOUSE (Shimizu et al., 2022), an AI drug discovery system that “illuminates” target compounds from a vast chemical space. This study was widely broadcasted in many media.
We are extending LIGHTHOUSE to develop a drug discovery platform that incorporates the laws of physiochemistry. We have begun to take on the challenge of designing not only small-molecule drugs but also more complex molecules such as nanobodies.
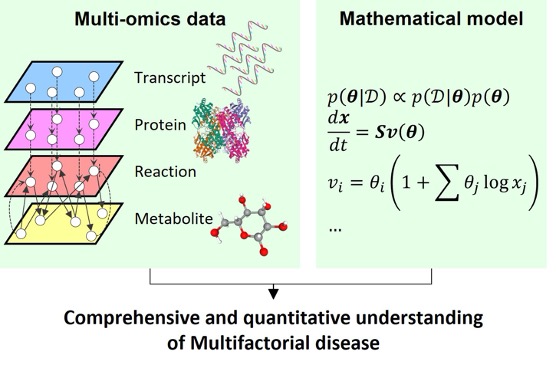
Understanding Multifactorial Diseases by Integrating Multi-omic Data
The cause of multifactorial diseases such as diabetes and cancer cannot be attributed to a single gene or molecule. Therefore, we aim comprehensive and quantitative understanding of the pathogenesis of multifactorial diseases by integrating multi-omic data, including metabolites, proteins, and RNA using mathematical analysis (Kodama, Oshikawa, Shimizu et al., 2020; Uematsu, Ohno, et al., iScience, 2022)。
AI-based stratification for personalized medicine (clinical informatics)
Based on our expertise as physicians, we are developing analytical methods and tools to address issues unique to healthcare data and conducting research for personalized medicine. For example, by using AI to learn breast cancer gene expression data, we developed a method to stratify stages based on only 23 genes (Shimizu et al., 2019), which was broadcasted in NHK news and Nikkei newspaper.
Others
We are doing a variety of other research in addition to the above three. Please contact us for more information.
Publications
- Sakuma T, Mukai Y, Yamaguchi A, Suganuma Y, Okamoto K, Furugen A, Narumi K, Ishikawa S, Saito Y,Kobayashi M. Monocarboxylate Transporters 1 and 2 Are Responsible for L-Lactate Uptake in Differentiated Human Neuroblastoma SH-SY5Y Cells. Biol. Pharm. Bull. 47(4):764-770, 2024
- Kasai S , Shiomi A, Shimizu H, Aoba M, Kinugasa Y, Miura T, Uehara K, Watanabe J, Kawai K, Ajioka Y.Risk factors and development of machine learning diagnostic models for lateral lymph node metastasis in rectal cancer: a multicentre study. BJS Open, 2024
- Konno Y, Uriu K, Chikata T, Takada T, Kurita JI, Ueda MT, Islam S, Yang Tan BJ, Ito J, Aso H, Kumata R, Williamson C, Iwami S, Takiguchi M, Nishimura Y, Morita E, Satou Y, Nakagawa S, Koyanagi Y, Sato K. Two-step evolution of HIV-1 budding system leading to pandemic in the human population. Cell Rep., 43(2), 113697, 2024
- Bai Y, Morita K, Kokaji T, Hatano A, Ohno S, Egami R, Pan Y, Li D, Yugi K, Uematsu S, Inoue H, Inaba Y, Suzuki Y, Matsumoto M, Izumi Y, Bamba T, Hirayama A, Soga T, Kuroda S. iScience, 27(3):109121, 2024
- 岡田随象 企画、川原田 明徳, 大谷 悠喜, 清水 秀幸分担著書、医学のあゆみ増刊号「遺伝統計学の新潮流 ─新規創薬・個別化医療への挑戦」, 医歯薬出版
- Oka T, Higa T, Sugahara O, Koga D, Nakayama S, Nakayama KI. Ablation of p57+ quiescent cancer stem cells suppresses recurrence after chemotherapy of intestinal tumors. Cancer Res., 2023
- Kodama M, Toyokawa G, Sugahara O, Sugiyama S, Haratake N, Yamada Y, Wada R, Takamori S, Shimokawa M, Takenaka T, Tagawa T, Kittaka H, Tsuruda T, Tanaka K, Komatsu Y, Nakata K, Imado Y, Yamazaki K, Okamoto I, Oda Y, Takahashi M, Izumi Y, Bamba T, Shimizu H, Yoshizumi T, Nakayama KI. Modulation of host glutamine anabolism enhances the sensitivity of small cell lung cancer to chemotherapy. Cell Rep., 42(8):112899, 2023
- Aso H, Ito J, Ozaki H, Kashima Y, Suzuki Y, Koyanagi Y, Sato K. Single-cell transcriptome analysis illuminating the characteristics of species-specific innate immune responses against viral infections. GigaScience, 12, 2023
- Funasaki S, Hatano A, Nakatsumi H, Koga D, Sugahara O, Yumimoto K, Baba M, Matsumoto M, Nakayama KI. A stepwise and digital pattern of RSK phosphorylation determines the outcome of thymic selection. iScience 26 (9), 107552, 2023
- Maehara H, Kokaji T, Hatano A, Suzuki Y, Matsumoto M, Nakayama KI, Egami R, Tsuchiya T, Ozaki H, Morita K, Shirai M, Li D, Terakawa A, Uematsu S, Hironaka K, Ohno S, Kubota H, Araki H, Miura F, Ito T, Kuroda S. DNA hypomethylation characterizes genes encoding tissue-dominant functional proteins in liver and skeletal muscle. Sci. Rep., 2023
- 大澤 毅 編、大野 聡 分担著書、実験医学増刊「マルチオミクス データ駆動時代の疾患研究」. 羊土社 2023
- 「一家に一枚 ウイルス」製作チーム (小嶋将平、渡士幸一、渡辺登喜子、堀江真行、麻生啓文、多賀佳、川久保修介、塩野谷果歩) 製作監修. 学習資料「一家に1枚 ウイルス」 文部科学省 2023
- Hozumi H, Shimizu H. Bayesian network enables interpretable and state-of-the-art prediction of immunotherapy responses in cancer patients. PNAS Nexus. in press
- Fujinuma S, Nakatsumi H, Shimizu H, Sugiyama S, Harada A, Goya T, Tanaka M, Kohjima M, Takahashi M, Izumi Y, Yagi M, Kang D, Kaneko M, Shigeta M, Bamba T, Ohkawa Y, Nakayama KI. FOXK1 promotes nonalcoholic fatty liver disease by mediating mTORC1-dependent inhibition of hepatic fatty acid oxidation. Cell Rep. in press.
- 清水 秀幸 編著. 実験医学別冊 Pythonで実践 生命科学データの機械学習. 羊土社 2023
- Uematsu S, Ohno S, Tanaka KY, Hatano A, Kokaji T, Ito Y, Kubota H, Hironaka K, Suzuki Y, Matsumoto M, Nakayama KI, Hirayama A, Soga T, Kuroda S. Multi-omics-based label-free metabolic flux inference reveals obesity-associated dysregulatory mechanisms in liver glucose metabolism. iScience, 25(2), 103787. (2022)
- Ohno S, Uematsu S, Kuroda S. Quantitative metabolic fluxes regulated by trans-omic networks. Biochem. J. 479(6), 787–804, 2022
- Kokaji T, Eto M, Hatano A, Yugi K, Morita K, Ohno S, Fujii M, Hironaka K, Ito Y, Egami R, Uematsu S, Terakawa A, Pan Y, Maehara H, Li D, Bai Y, Tsuchiya T, Ozaki H, Inoue H, Kubota H, Suzuki Y, Hirayama A, Soga T, Kuroda S. In vivo transomic analyses of glucose-responsive metabolism in skeletal muscle reveal core differences between the healthy and obese states. Sci. Rep. 12(1), 1–19, 2022
- Aso H, Ito J, Ozaki H, Kashima Y, Suzuki Y, Koyanagi Y, Sato K. Single-cell transcriptome analysis illuminating the characteristics of species-specific innate immune responses against viral infections. bioRxiv, 2022
- Mise S, Matsumoto A, Shimada K, Hosaka T, Takahashi M, Ichihara K, Shimizu H, Shiraishi C, Saito D, Suyama M, Yasuda T, Ide T, Izumi Y, Bamba T, Kimura-Someya T, Shirouzu T, Miyata H, Ikawa M, and Nakayama KI. Kastor and Polluks polypeptides encoded by a single gene locus cooperatively regulate VDAC and spermatogenesis. Nature Commun. 2022
- Shimizu H, Kodama M, Matsumoto M, Orba Y, Sasaki Y, Sato A, Sawa H, Nakayama KI. LIGHTHOUSE illuminates therapeutics for a variety of diseases including COVID-19. iScience 2022
- Habara M, Sato Y, Goshima T, Sakurai M, Imai H, Shimizu H, Katayama Y, Hanaki S, Masaki T, Morimoto M, Nishikawa S, Toyama T, Shimada M. PNAS 2022
- Terakawa A, Hu Y, Kokaji T, Yugi K, Morita K, Ohno S, Pan Y, Bai Y, Parkhitko AA, Ni X, Asara JM, Bulyk ML, Perrimon N, Kuroda S. iScience 2022
- Aso H, Nagaoka S, Kawakami, Ito J, Islam S, Tan BJY, Nakaoka S, Ashizaki K, Shirogushi K, Suzuki Y, Satou Y, Koyanagi Y, Sato K. Multiomics investigation revealing comprehensive characteristics of HIV-1-infected cells in vivo. Cell Rep., 32 (2), 107887, 2020
- Kosugi Y, Uriu K, Suzuki N, Yamamoto K, Nagaoka S, Kimura I, Konno Y, Aso H, Willett BJ, Kobayashi T, Koyanagi Y, Ueda MT, Ito J, Sato K. A comprehensive investigation on the interplay between feline APOBEC3Z3 proteins and feline immunodeficiency virus Vif proteins. J. Virol., 95(13):e0017821, 2021
- Ichihara K, Matsumoto A, Nishida H, Kito Y, Shimizu H, Shichino Y, Iwasaki S, Imami K, Ishihama Y, Nakayama KI. Combinatorial analysis of translation dynamics reveals eIF2 dependence of translation initiation at near-cognate codons. Nucleic Acids Res. 2021
- Shimizu H, Nakayama KI. A universal molecular prognostic score for gastrointestinal tumors. NPJ Genom. Med. 2021
- Egami R, Kokaji T, Hatano A, Yugi K, Eto M, Morita K, Ohno S, Fujii M, Hironaka K, Uematsu S, Terakawa A, Bai Y, Pan Y, Tsuchiya T, Ozaki H, Inoue H, Uda S, Kubota H, Suzuki Y, Matsumoto M, Nakayama KI, Hirayama A, Soga T, Kuroda S. iScience 2021
- Hwang YS, Suzuki S, Saita Y, Ito J, Sakata Y, Aso H, Sato K, Hermann BP, Sasaki K. Reconstitution of prospermatogonial specification in vitro from human induced pluripotent stem cells. Nature Commun.,11(1), 5656, 2020
- Nakano Y, Keisuke Y, Ueda MT, Soper A, Konno Y, Kimura I, Uriu K, Kumata R, Aso H, Misawa N, Nagaoka S, Shimizu S, Mitsumune K, Kosugi Y, Juarez-Fernandez G, Ito J, Nakagawa S, Ikeda T, Koyanagi Y, Harris RS, Sato K. A role for gorilla APOBEC3G in shaping lentivirus evolution including transmission to humans. PLoS Pathog., 16(9), e1008812, 2020
- Onoyama I, Nakayama S, Shimizu H, Nakayama KI. Loss of Fbxw7 impairs development of and induces heterogeneous tumor formation in the mouse mammary gland. Cancer Res. 2020
- Shimizu H, Nakayama KI. Artificial intelligence in oncology. Cancer Sci. 2020
- Yamauchi Y, Nita A, Nishiyama M, Muto Y, Shimizu H, Nakatsumi H, Nakayama KI. Skp2 contributes to cell cycle progression in trophoblast stem cells and to placental development. Genes Cells. 2020
- Oshikawa K, Matsumoto M, Kodama M, Shimizu H, Nakayama KI. A fail-safe system to prevent oncogenesis by senescence is targeted by SV40 small T antigen. Oncogene 2020
- Kodama M, Oshikawa K, Shimizu H, Yoshioka S, Takahashi M, Izumi Y, Bamba T, Tateishi C, Tomonaga T, Matsumoto M, Nakayama KI. A shift in glutamine nitrogen metabolism contributes to malignant progression of cancer. Nature Commun. 2020
- Kokaji T, Hatano A, Ito Y, Yugi K, Eto M, Morita K, Ohno S, Fujii M, Hironaka K, Egami R, Terakawa A, Tsuchiya T, Ozaki H, Inoue H, Uda S, Kubota H, Suzuki Y, Ikeda K, Arita M, Matsumoto M, Nakayama K.I, Hirayama A, Soga T, Kuroda S. Science Signaling 2020
- Ohno S, Quek L-E, Krycer JR, Yugi K, Hirayama A, Ikeda S, Shoji F, Suzuki K, Soga T, James DE, Kuroda S. iScience 2020
- Quek L-E, Krycer JR, Ohno S, Yugi K, Fazakerley DJ, Scalzo R, Elkington SD, Dai Z, Hirayama A, Ikeda S, Shoji F, Suzuki K, Locasale JW, Soga T, James DE, Kuroda S. iScience 2020
- Aso H, Ito J, Koyanagi Y, Sato K. Comparative Description of the Expression Profile of Interferon- stimulated Genes in Multiple Cell Lineages Targeted by HIV-1 Infection. Front. Microbiol., 10, 429, 2019
- Shimizu H, Nakayama KI. A 23 gene-based molecular prognostic score precisely predicts overall survival of breast cancer patients. EBioMedicine 2019
- Muto Y, Moroishi T, Ichihara K, Nishiyama M, Shimizu H, Eguchi H, Moriya K, Koike K, Mimori K, Mori M, Katayama Y, Nakayama KI. Disruption of FBXL5-mediated cellular iron homeostasis promotes liver carcinogenesis. J. Exp. Med. 2019
- Shimizu H, Takeishi S, Nakatsumi H, Nakayama KI. Prevention of cancer dormancy by Fbxw7 ablation eradicates disseminated tumor cells. JCI Insight 2019
- Krycer JR, Yugi K, Hirayama A, Fazakerley DJ, Quek L-E, Scalzo R, Ohno S, Hodson MP, Ikeda S, Shoji F, Suzuki K, Domanova W, Parker LB, Nelson ME, Humphrey SJ, Turner N, Hoehn KL, Cooney GJ, Soga T, Kuroda S, James DE. Cell Reports 2017
- Sano T, Kawata K, Ohno S, Yugi K, Kakuda H, Kubota H, Uda S, Fujii M, Kunida K, Hoshino D, Hatano A, Ito Y, Sato M, Suzuki Y, Kuroda S. Science Signaling 2016
- Tokuyama K, Ohno S, Yoshikawa K, Hirasawa T, Tanaka S, Furusawa C, Shimizu H. Microb. Cell Fact. 2014
- Nakashima N, Ohno S, Yoshikawa K, Shimizu H, Tamura T. Appl. Environ. Microbiol. 2014
- Ohno S, Shimizu H, Furusawa C. Bioinformatics 2014
- Ohno S, Furusawa C, Shimizu H. J. Biosci. Bioeng. 2013
- Saiki Y, Yoshino Y, Fujimura H, Manabe T, Kudo Y, Shimada M, Mano N, Nakano T, Lee Y, Shimizu S, Oba S, Fujiwara S, Shimizu H, Chen N, Nezhad ZK, Jin G, Fukushige S, Sunamura M, Ishida M, Motoi F, Egawa S, Unno M, Horii A. DCK is frequently inactivated in acquired gemcitabine-resistant human cancer cells. Biochem. Biophys. Res. Commun. 2012
- Shimizu H, Horii A, Sunamura M, Motoi F, Egawa S, Unno M, Fukushige S. Identification of epigenetically silenced genes in human pancreatic cancer by a novel method “microarray coupled with methyl-CpG targeted transcriptional activation” (MeTA-array). Biochem. Biophys. Res. Commun. 2011

Department of Integrated Analytics

Department of AI Technology Development

Department of Biostatistics


